Introduction
The sbt software is an R package (R Core Team 2024) that
contains the CCSBT operating model (OM) coded using RTMB Kristensen (2024). This page provides an
example of using the sbt model.
Load inputs
The sbt RTMB model is loaded along with several R
functions using library(sbt). The kableExtra
package is used for generating tables of inputs. The
tidyverse package is used for data manipulation and
plotting. The code theme_set(theme_bw()) alters the plot
aesthetics.
# remotes::install_github("janoleko/RTMBdist")
# remotes::install_github("andrjohns/StanEstimators")
# remotes::install_github("Cole-Monnahan-NOAA/adnuts")
library(kableExtra)
library(tidyverse)
library(sbt)
theme_set(theme_bw())Below are example inputs that are loaded from the sbt
package. Normally the user would import these inputs (e.g., from
.csv files using the read_csv function).
# names(data_csv1)
# names(data_par1)
# names(data_labrep1)The data list is defined below and then the get_data
function is used to set up some additional inputs that are required
before the data list can be passed to MakeADFun.
data <- list(
last_yr = 2022, age_increase_M = 25, length_m50 = 150, length_m95 = 180,
catch_UR_on = 0, catch_surf_case = 1, catch_LL1_case = 1,
scenarios_surf = data_csv1$scenarios_surface,
scenarios_LL1 = data_csv1$scenarios_LL1,
removal_switch_f = c(0, 0, 0, 1, 0, 0), # 0=harvest rate, 1=direct removals
sel_min_age_f = c(2, 2, 2, 8, 6, 0, 2),
sel_max_age_f = c(17, 9, 17, 22, 25, 7, 17),
sel_end_f = c(1, 0, 1, 1, 1, 0, 1),
sel_LL1_yrs = c(1952, 1957, 1961, 1965, 1969, 1973, 1977, 1981, 1985, 1989,
1993, 1997, 2001, 2006, 2007, 2008, 2011, 2014, 2017, 2020),
sel_LL2_yrs = c(1969, 2001, 2005, 2008, 2011, 2014, 2017, 2020),
sel_LL3_yrs = c(1954, 1961, 1965, 1969, 1970, 1971, 2005, 2006, 2007),
sel_LL4_yrs = c(1953),
sel_Ind_yrs = c(1976, 1995, 1997, 1999, 2002, 2004, 2006, 2008, 2010, 2012,
2013, 2014, 2015, 2016, 2017, 2018, 2019, 2020, 2021, 2022),
sel_Aus_yrs = c(1952, 1969, 1973, 1977, 1981, 1985, 1989, 1993, 1997, 1998,
1999, 2000, 2001, 2002, 2003, 2004, 2005, 2006, 2007, 2008,
2009, 2010, 2011, 2012, 2013, 2014, 2015, 2016, 2017, 2018,
2019, 2020, 2021, 2022),
sel_CPUE_yrs = c(1969, 1973, 1977, 1981, 1985, 1989, 1993, 1997, 2001, 2006,
2007, 2008, 2011, 2014, 2017, 2020),
af_switch = 9,
lf_switch = 9, lf_minbin = c(1, 1, 1, 11),
cpue_switch = 1, cpue_a1 = 5, cpue_a2 = 17,
troll_switch = 1,
aerial_switch = 4, aerial_tau = 0.3,
tag_switch = 1, tag_var_factor = 1.82,
hsp_switch = 1, hsp_false_negative = 0.7467647,
pop_switch = 1,
gt_switch = 1
)
data <- get_data(data_in = data)
names(data)
#> [1] "last_yr" "age_increase_M" "length_m50"
#> [4] "length_m95" "removal_switch_f" "sel_min_age_f"
#> [7] "sel_max_age_f" "sel_end_f" "af_switch"
#> [10] "lf_switch" "lf_minbin" "cpue_switch"
#> [13] "cpue_a1" "cpue_a2" "troll_switch"
#> [16] "aerial_switch" "aerial_tau" "tag_switch"
#> [19] "tag_var_factor" "hsp_switch" "hsp_false_negative"
#> [22] "pop_switch" "gt_switch" "first_yr"
#> [25] "n_year" "n_season" "min_age"
#> [28] "max_age" "n_age" "n_length"
#> [31] "n_fishery" "age_a" "length_mu_ysa"
#> [34] "length_sd_a" "weight_fya" "first_yr_catch"
#> [37] "first_yr_catch_f" "n_catch" "catch_year"
#> [40] "catch_obs_ysf" "sel_change_year_fy" "pop_obs"
#> [43] "paly" "hsp_obs" "gt_obs"
#> [46] "gt_nscan" "gt_nrec" "aerial_years"
#> [49] "aerial_obs" "aerial_cv" "aerial_cov"
#> [52] "troll_years" "troll_obs" "troll_sd"
#> [55] "cpue_years" "cpue_obs" "cpue_sd"
#> [58] "af_min_age" "af_max_age" "n_af"
#> [61] "af_year" "af_fishery" "af_obs"
#> [64] "af_n" "dl_yal" "alk_ysal"
#> [67] "lf_year" "lf_fishery" "lf_season"
#> [70] "lf_obs" "lf_n" "lf_slices"
#> [73] "af_sliced" "af_sliced_ysfa" "cpue_lfs"
#> [76] "cpue_n" "tag_shed_immediate" "tag_shed_continuous"
#> [79] "tag_rep_rates_ya" "tag_rel_min_age" "tag_rel_max_age"
#> [82] "tag_release_cta" "tag_recap_ctaa" "tag_recap_max_age"
#> [85] "min_K" "n_K" "n_T"
#> [88] "n_I" "n_J"Input tables
Below are examples of some of the model inputs in tabular form.
Mean length (cm) by year, season, and age. Only a subset of years and ages 1-10 are shown:
kbl(bind_rows(head(length_mean), tail(length_mean))[,1:13]) %>%
add_header_above(c(" " = 2, "Age" = 11))| Year | Season | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1931 | 1 | 45 | 57.1 | 74.1 | 88.8 | 101.8 | 113.9 | 124.3 | 133.3 | 141.0 | 147.6 | 153.3 |
| 1932 | 1 | 45 | 57.1 | 74.1 | 88.8 | 101.8 | 113.9 | 124.3 | 133.3 | 141.0 | 147.6 | 153.3 |
| 1933 | 1 | 45 | 57.1 | 74.1 | 88.8 | 101.8 | 113.9 | 124.3 | 133.3 | 141.0 | 147.6 | 153.3 |
| 1934 | 1 | 45 | 57.1 | 74.1 | 88.8 | 101.8 | 113.9 | 124.3 | 133.3 | 141.0 | 147.6 | 153.3 |
| 1935 | 1 | 45 | 57.1 | 74.1 | 88.8 | 101.8 | 113.9 | 124.3 | 133.3 | 141.0 | 147.6 | 153.3 |
| 1936 | 1 | 45 | 57.1 | 74.1 | 88.8 | 101.8 | 113.9 | 124.3 | 133.3 | 141.0 | 147.6 | 153.3 |
| 2017 | 2 | 50 | 68.9 | 91.6 | 105.5 | 117.3 | 127.2 | 135.6 | 142.7 | 148.7 | 153.8 | 158.1 |
| 2018 | 2 | 50 | 68.9 | 91.6 | 105.5 | 117.3 | 127.2 | 135.6 | 142.7 | 148.7 | 153.8 | 158.1 |
| 2019 | 2 | 50 | 68.9 | 91.6 | 105.5 | 117.3 | 127.2 | 135.6 | 142.7 | 148.7 | 153.8 | 158.1 |
| 2020 | 2 | 50 | 68.9 | 91.6 | 105.5 | 117.3 | 127.2 | 135.6 | 142.7 | 148.7 | 153.8 | 158.1 |
| 2021 | 2 | 50 | 68.9 | 91.6 | 105.5 | 117.3 | 127.2 | 135.6 | 142.7 | 148.7 | 153.8 | 158.1 |
| 2022 | 2 | 50 | 68.9 | 91.6 | 105.5 | 117.3 | 127.2 | 135.6 | 142.7 | 148.7 | 153.8 | 158.1 |
The parent offspring pair (POP) data set:
| Cohort | CaptureYear | CaptureCov | CaptureSwitch | NPOPS | Comps |
|---|---|---|---|---|---|
| 2007 | 2012 | 15 | 1 | 0 | 1315 |
| 2008 | 2012 | 15 | 1 | 0 | 963 |
| 2009 | 2012 | 15 | 1 | 0 | 876 |
| 2010 | 2012 | 15 | 1 | 0 | 903 |
| 2011 | 2012 | 15 | 1 | 0 | 899 |
| 2003 | 2013 | 15 | 1 | 0 | 1317 |
| 2004 | 2013 | 15 | 1 | 0 | 1325 |
| 2005 | 2013 | 15 | 1 | 0 | 1356 |
| 2006 | 2013 | 15 | 1 | 0 | 1347 |
| 2007 | 2013 | 15 | 1 | 0 | 1315 |
| 2008 | 2013 | 15 | 1 | 0 | 963 |
| 2009 | 2013 | 15 | 1 | 0 | 876 |
| 2010 | 2013 | 15 | 1 | 0 | 903 |
| 2011 | 2013 | 15 | 1 | 0 | 899 |
| 2012 | 2013 | 15 | 1 | 0 | 953 |
The unaccounted catch (tonnes) data set:
| Year | LL1 | LL2 | LL3 | LL4 | Indonesia | Australia |
|---|---|---|---|---|---|---|
| 2008 | 72 | 0 | 0 | 0 | 0 | 0 |
| 2009 | 152 | 0 | 0 | 0 | 0 | 0 |
| 2010 | 271 | 0 | 0 | 0 | 0 | 0 |
| 2011 | 151 | 0 | 0 | 0 | 0 | 0 |
| 2012 | 275 | 0 | 0 | 0 | 0 | 0 |
| 2013 | 432 | 0 | 0 | 0 | 0 | 0 |
| 2014 | 121 | 0 | 0 | 0 | 0 | 0 |
| 2015 | 326 | 0 | 0 | 0 | 0 | 0 |
| 2016 | 756 | 0 | 0 | 0 | 0 | 0 |
| 2017 | 984 | 0 | 0 | 0 | 0 | 0 |
| 2018 | 1511 | 0 | 0 | 0 | 0 | 0 |
| 2019 | 1155 | 0 | 0 | 0 | 0 | 0 |
| 2020 | 1160 | 0 | 0 | 0 | 0 | 0 |
| 2021 | 1160 | 0 | 0 | 0 | 0 | 0 |
| 2022 | 1160 | 0 | 0 | 0 | 0 | 0 |
Input plots
plot_length_at_age(data = data, years = c(1931, 2022))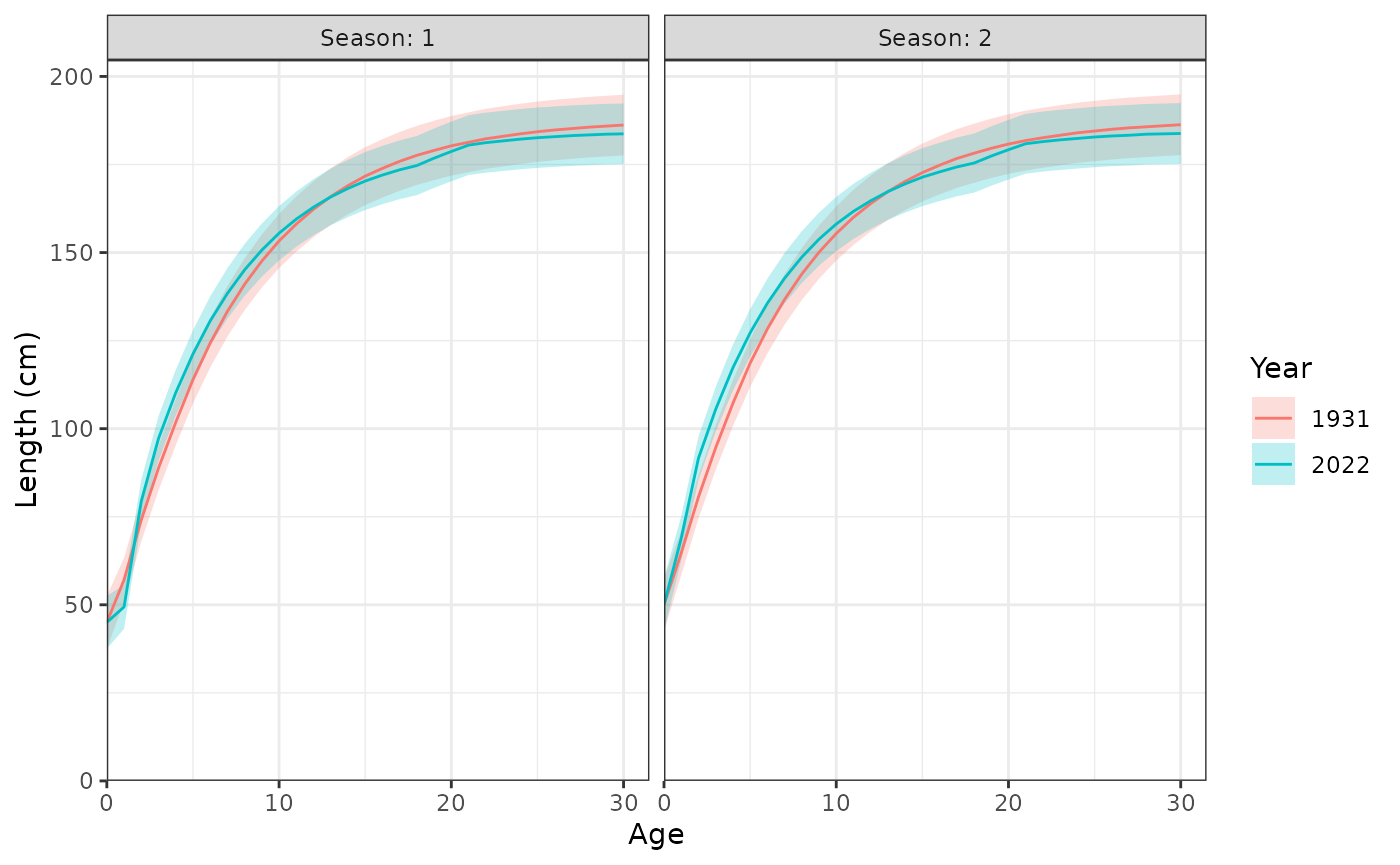
Length (cm) at age during each season.
plot_weight_at_age(data = data, years = c(1931, 2000, 2010, 2022))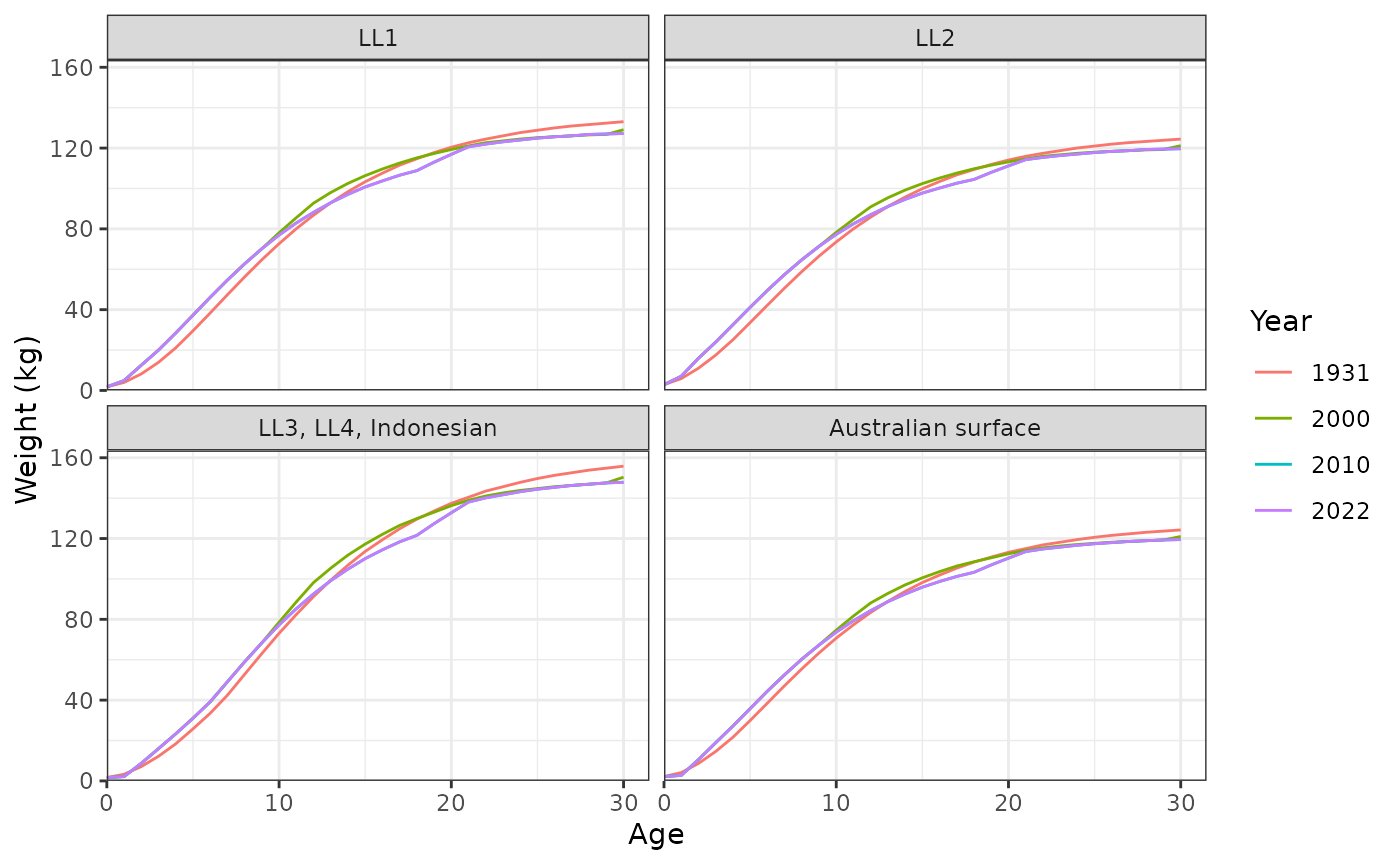
Weight (kg) at age for each fishery.
Model setup
Define the parameter list:
parameters <- get_parameters(data)
names(parameters)
#> [1] "par_log_B0" "par_log_psi" "par_log_m0"
#> [4] "par_log_m4" "par_log_m10" "par_log_m30"
#> [7] "par_log_h" "par_log_sigma_r" "par_log_cpue_q"
#> [10] "par_cpue_creep" "par_log_cpue_sigma" "par_log_cpue_omega"
#> [13] "par_log_aerial_tau" "par_log_aerial_sel" "par_log_troll_tau"
#> [16] "par_log_gt_q" "par_log_hsp_q" "par_log_tag_H_factor"
#> [19] "par_log_af_alpha" "par_log_lf_alpha" "par_sel_rho_y"
#> [22] "par_sel_rho_a" "par_log_sel_sigma" "par_log_sel_1"
#> [25] "par_log_sel_2" "par_log_sel_3" "par_log_sel_4"
#> [28] "par_log_sel_5" "par_log_sel_6" "par_log_sel_7"
#> [31] "par_rdev_y"There is a lot of flexibility in specifying priors now:
data$priors <- get_priors(parameters)
evaluate_priors(parameters, data$priors)
#> [1] 61.79457Use TMB’s Map option to turn parameters on/off:
map <- get_map(data, parameters)Using the data, the parameters, the
parameter map, and the model (sbt_model), the
AD object is created using TMBs MakeADFun function:
obj <- MakeADFun(func = cmb(sbt_model, data), parameters = parameters, map = map)List of parameters that are on:
unique(names(obj$par))
#> [1] "par_log_B0" "par_log_m4" "par_log_m30"
#> [4] "par_log_cpue_q" "par_log_troll_tau" "par_log_sel_1"
#> [7] "par_log_sel_2" "par_log_sel_3" "par_log_sel_5"
#> [10] "par_log_sel_6" "par_log_sel_7" "par_rdev_y"The objective function value:
obj$fn(obj$par)
#> [1] 24653.79Load the default parameter bounds:
bnd <- get_bounds(obj, parameters)Optimisation
Optimise using the nlminb function:
control <- list(eval.max = 10000, iter.max = 10000)
opt <- nlminb(start = obj$par, objective = obj$fn, gradient = obj$gr, hessian = obj$he,
lower = bnd$lower, upper = bnd$upper, control = control)
#> Warning in nlminb(start = obj$par, objective = obj$fn, gradient = obj$gr, :
#> NA/NaN function evaluation
#> Warning in nlminb(start = obj$par, objective = obj$fn, gradient = obj$gr, :
#> NA/NaN function evaluation
#> Warning in nlminb(start = obj$par, objective = obj$fn, gradient = obj$gr, :
#> NA/NaN function evaluation
#> Warning in nlminb(start = obj$par, objective = obj$fn, gradient = obj$gr, :
#> NA/NaN function evaluation
#> Warning in nlminb(start = obj$par, objective = obj$fn, gradient = obj$gr, :
#> NA/NaN function evaluation
opt <- nlminb(start = opt$par, objective = obj$fn, gradient = obj$gr, hessian = obj$he,
lower = bnd$lower, upper = bnd$upper, control = control)
# save(opt, file = "opt.rda")
# load("opt.rda")
# opt <- nlminb(start = opt$par, objective = obj$fn, gradient = obj$gr, hessian = obj$he,
# lower = bnd$lower, upper = bnd$upper, control = control)Check that all parameters are estimable:
# check_estimability(obj = obj)Try the analytical Hessian instead:
# he <- obj$he()
# he_inv <- solve(he)
# he_ch <- chol(he)
# ev <- eigen(he)
# range(ev$values)Calculate standard deviations of all model parameters, including non
linear functions of random effects and parameters specified through the
ADREPORT() macro from the user template:
Simulation
Simulation can be done for any data set in the model that is passed
through the RTMB function OBS. For example,
the CPUE series is set up using:
cpue_log_obs <- log(cpue_obs)
cpue_log_obs <- OBS(cpue_log_obs)
lp <- -dnorm(x = cpue_log_obs, mean = cpue_log_pred, sd = cpue_sig, log = TRUE)This allows a call to obj$simulate()$cpue_log_obs. For
example:
# obj$simulate()$cpue_log_obs
# obj$simulate()$troll_log_obs
# obj$simulate()$aerial_log_obs
# obj$simulate()$gt_nrec
# obj$simulate()$hsp_nK
# obj$simulate()$pop_nP
plot(log(data$cpue_obs), col = 2)
for (i in 1:10) lines(obj$simulate()$cpue_log_obs)
Data sets for which simulation is available include:
- cpue_log_obs
- troll_log_obs
- aerial_log_obs
- gt_nrec
- hsp_nK
- pop_nP
But not:
- af_obs
- lf_obs
- cpue_lfs
Simulation is also required for calculating OSA residuals and to use
the checkConsistency function. Unfortunately, the
RTMB function checkConsistency does not work
yet because not all data types are defined using densities that have
simulation support within the model (i.e., the AFs and LFs).
# chk <- checkConsistency(obj = obj, hessian = TRUE, estimate = TRUE, n = 100, observation.name = "cpue_log_obs")
# chk
# s <- summary(chk)
# s
# s$marginal$p.valuePlot outputs
Model fits
plot_cpue(data = data, object = obj, nsim = 10)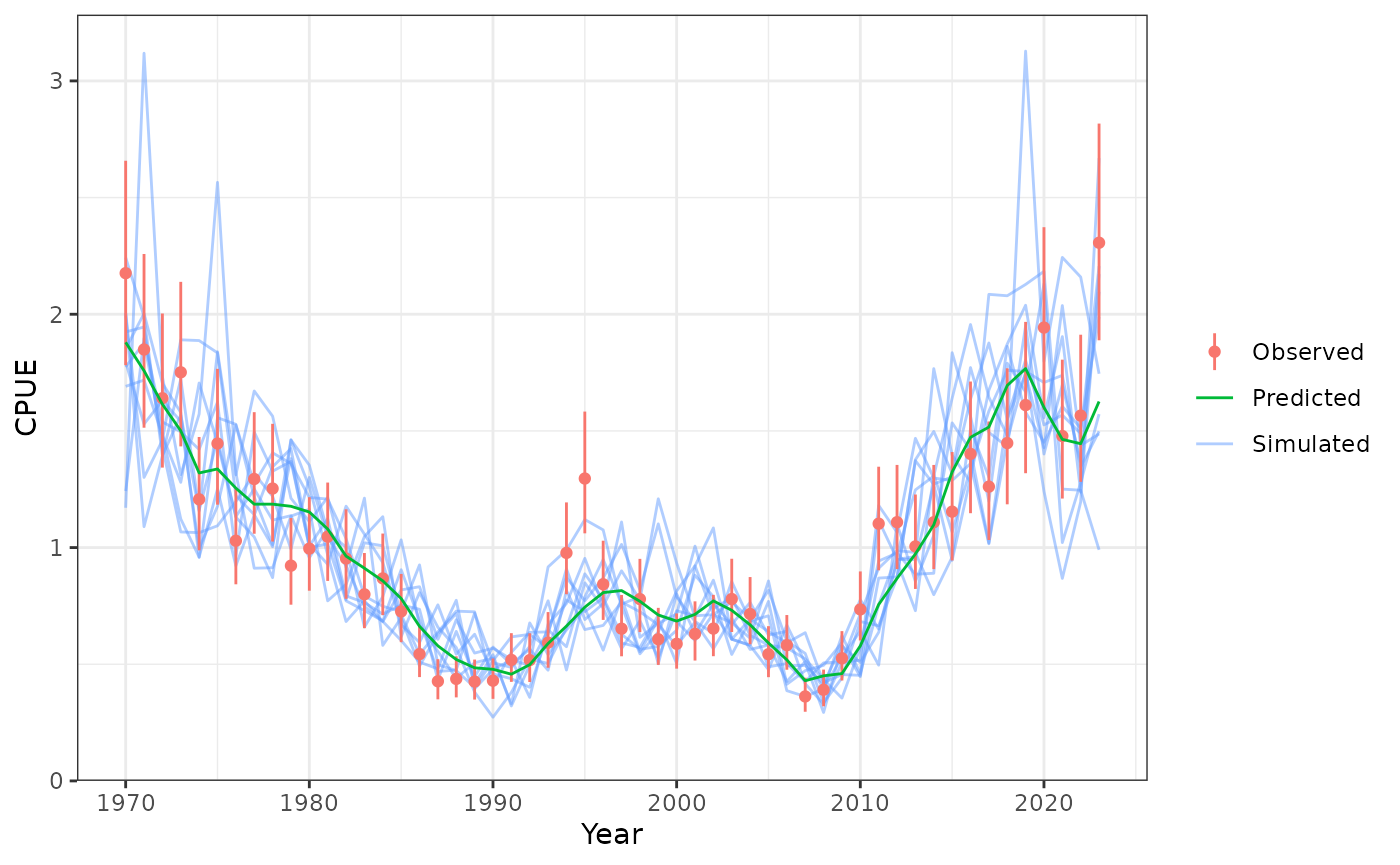
Model fits to CPUE.
plot_aerial_survey(data = data, object = obj, nsim = 10)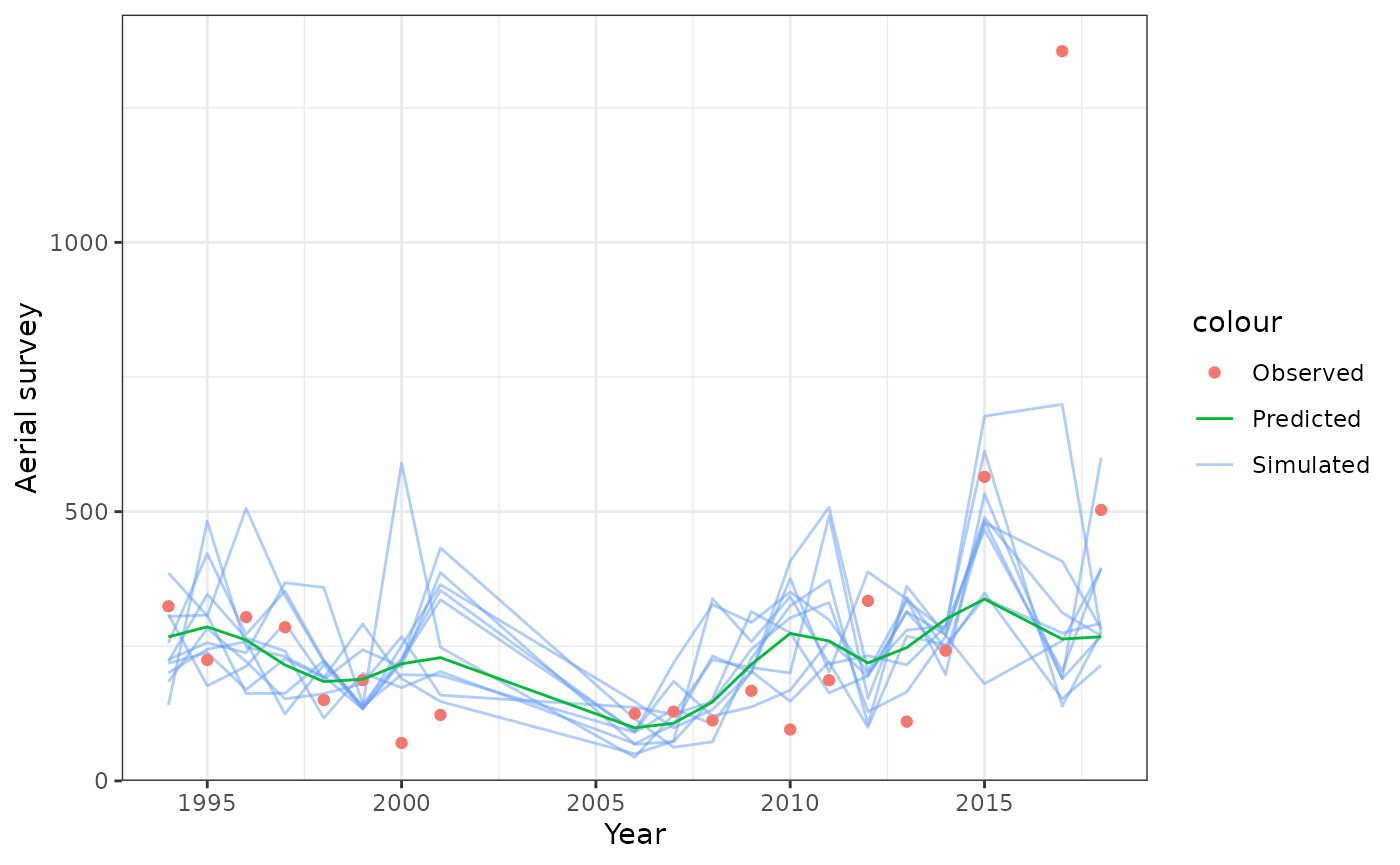
Model fits to aerial surveys.
plot_lf(data = data, object = obj, fishery = "LL1")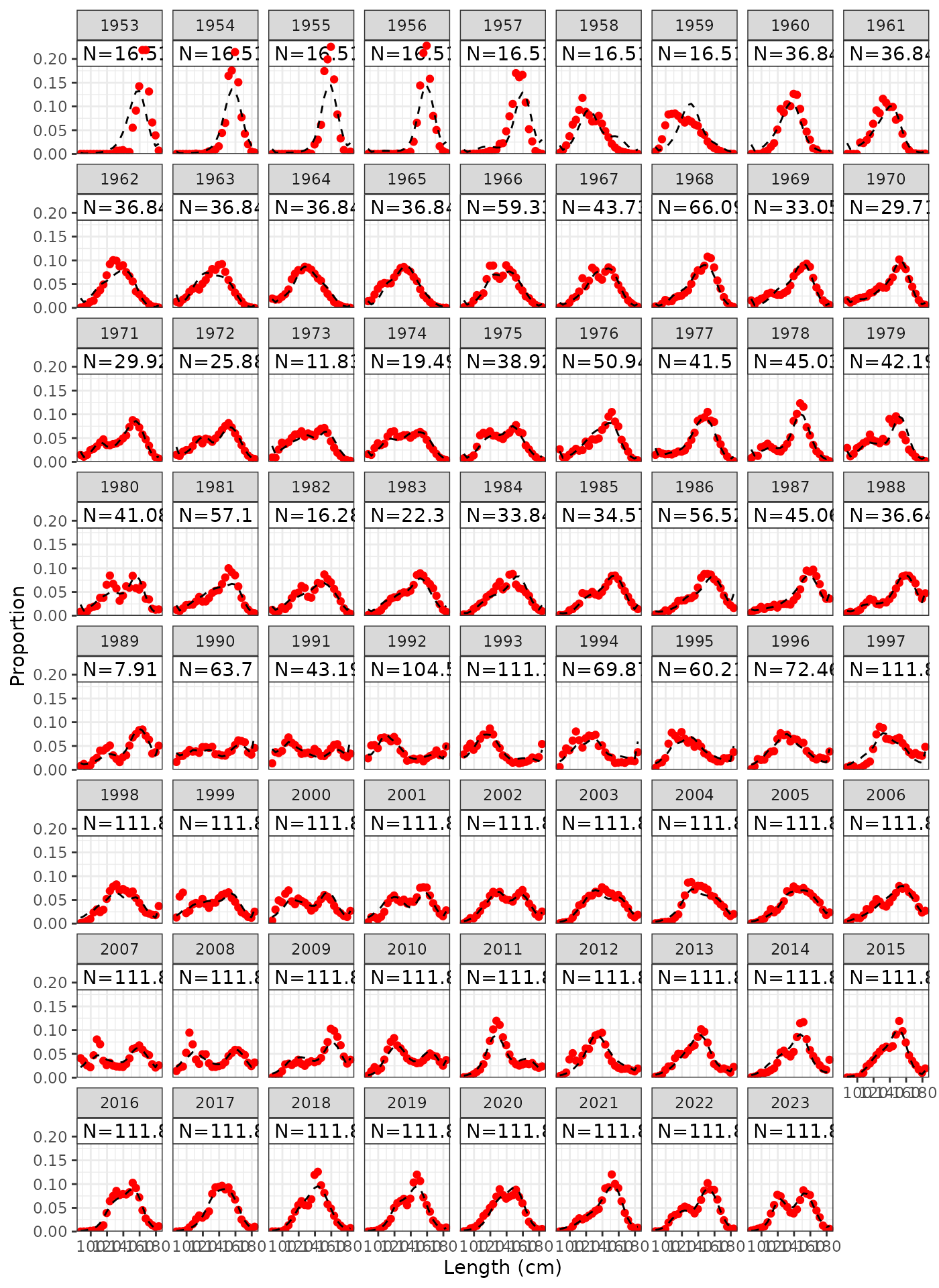
Model fits to LL1 length frequencies.
plot_lf(data = data, object = obj, fishery = "LL2")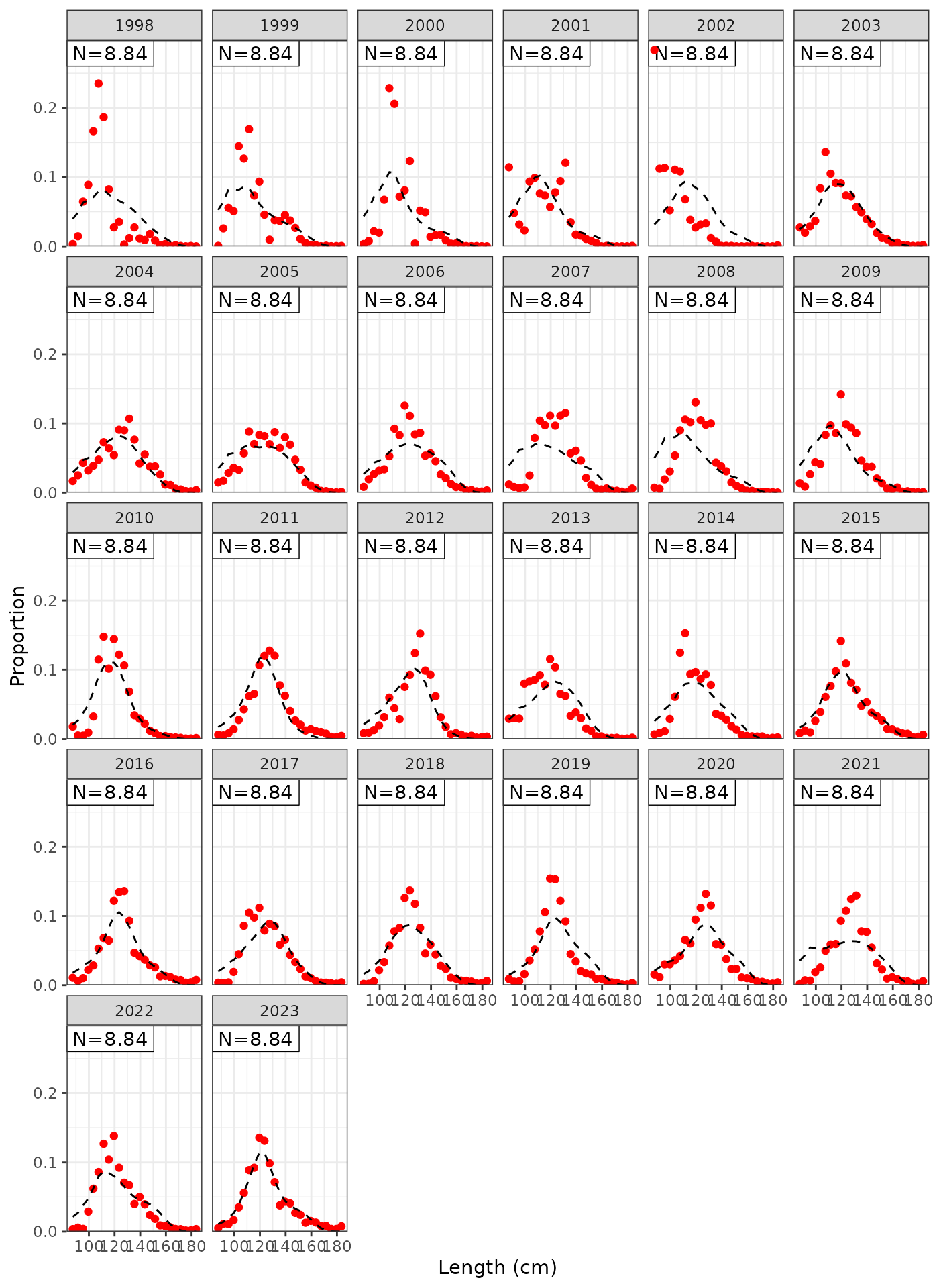
Model fits to LL2 length frequencies.
plot_lf(data = data, object = obj, fishery = "LL3")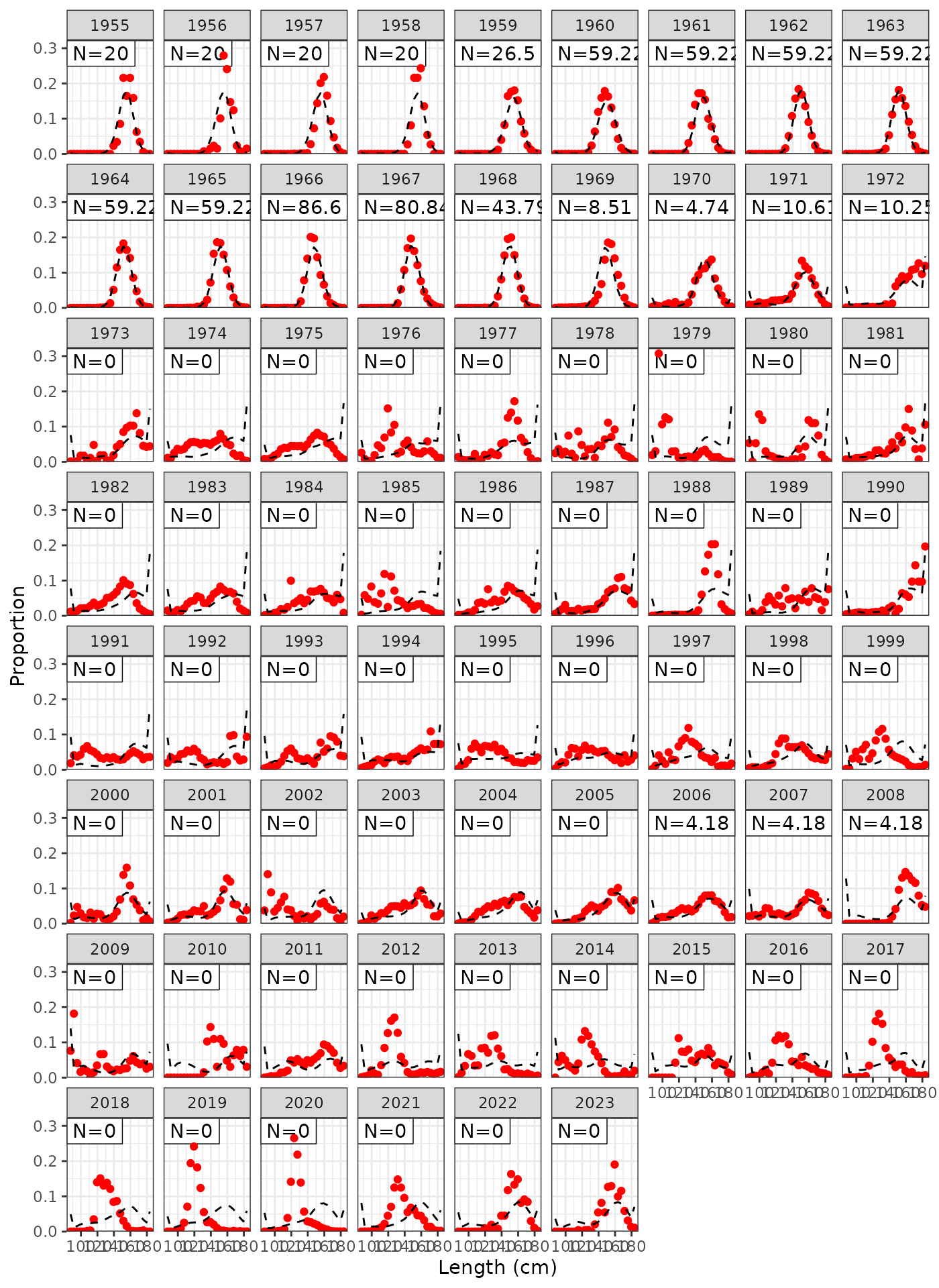
Model fits to LL3 length frequencies.
plot_lf(data = data, object = obj, fishery = "LL4")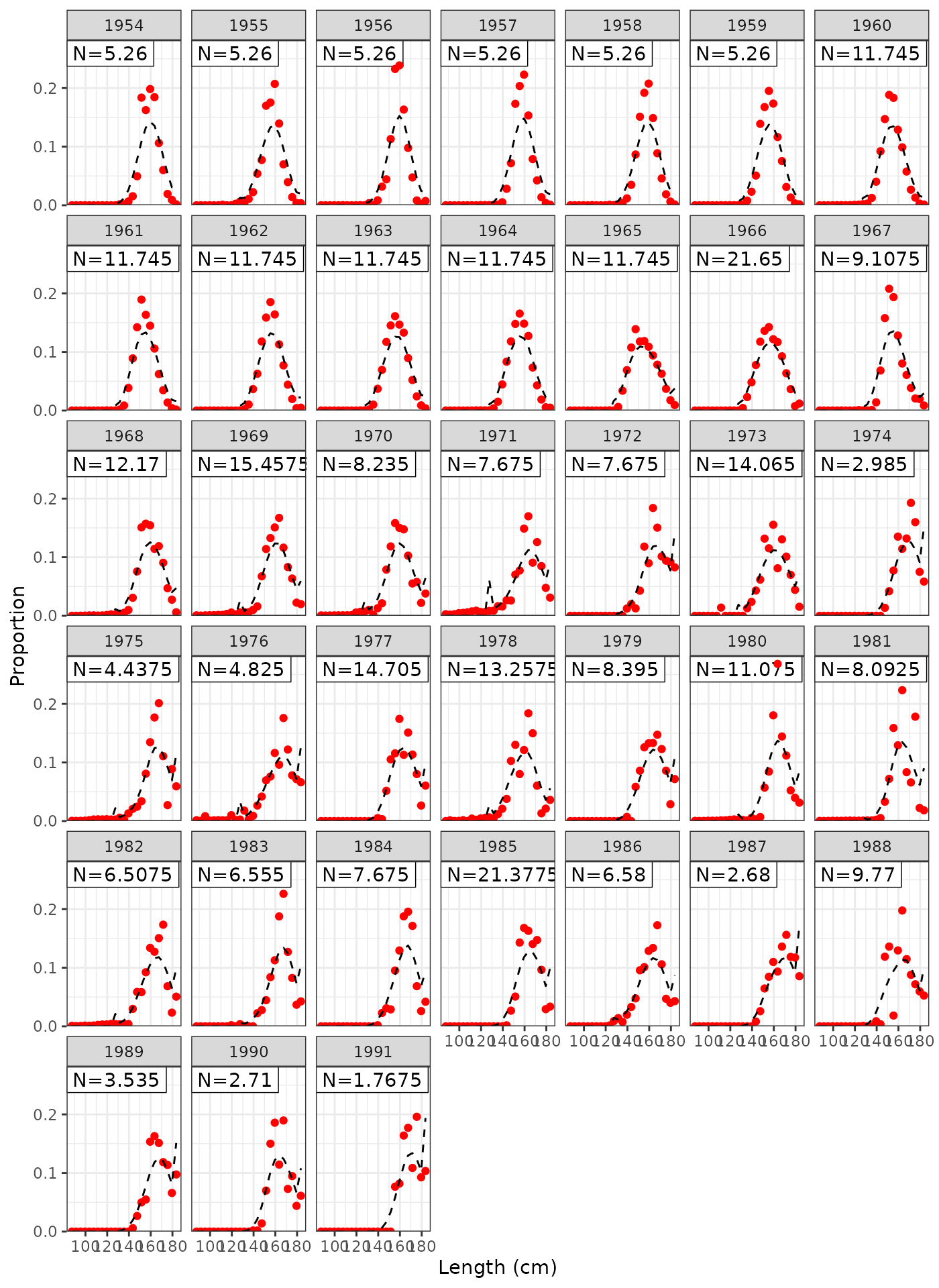
Model fits to LL4 length frequencies.
plot_af(data = data, object = obj, fishery = "Indonesian")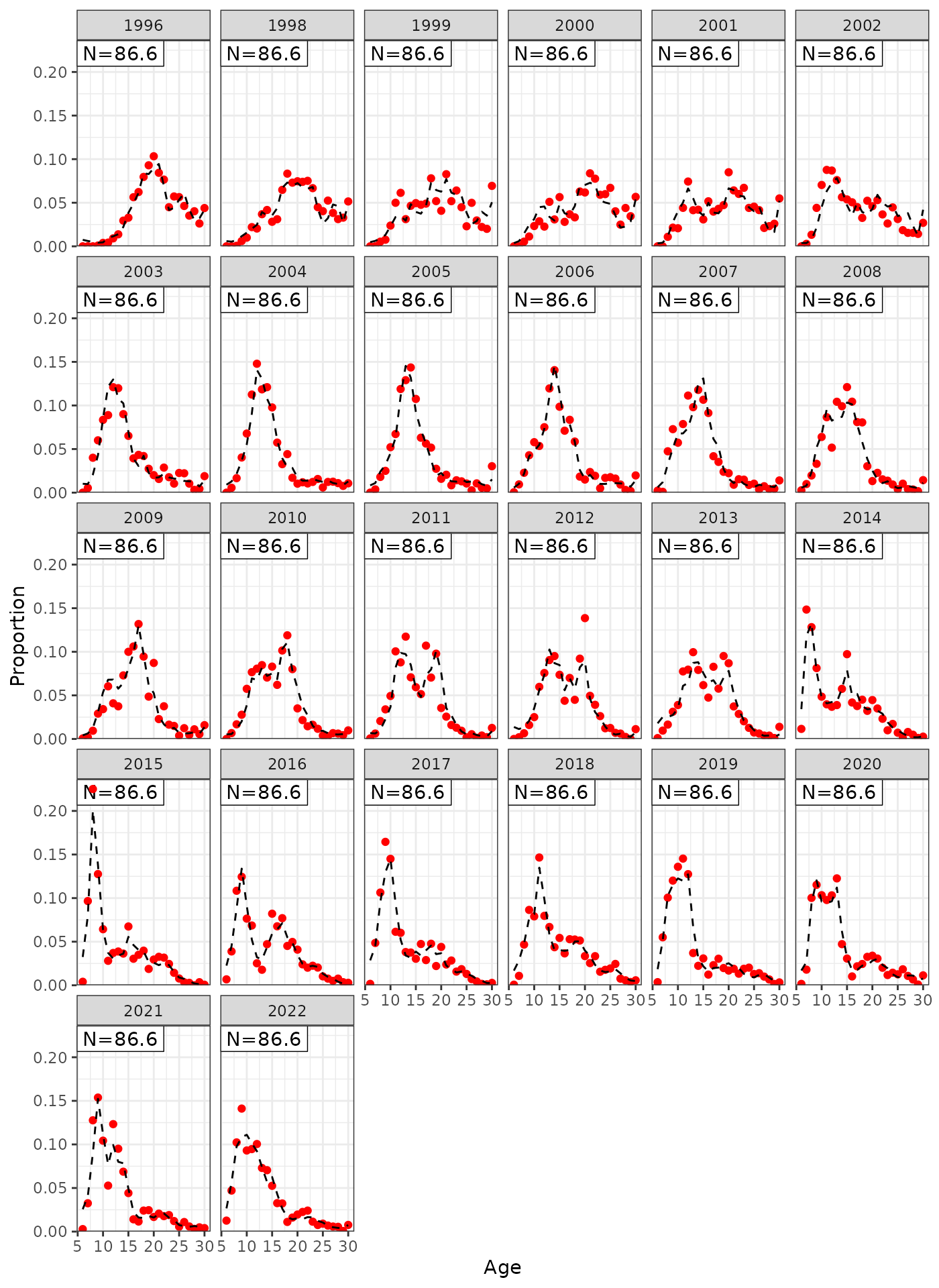
Model fits to Indonesian age compositions.
plot_af(data = data, object = obj, fishery = "Australian")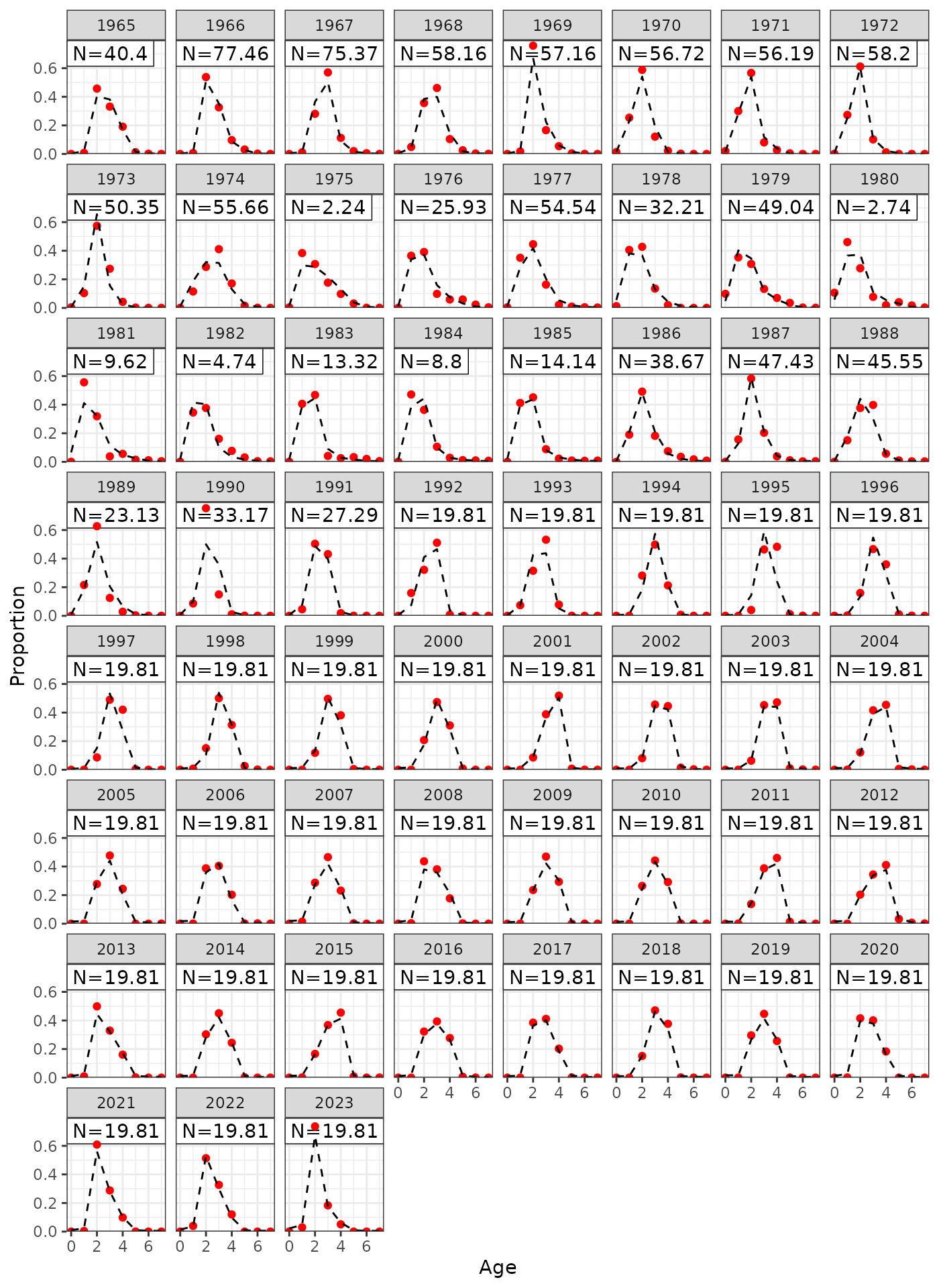
Model fits to Australian age compositions.
I think that not all of the catch is being taken for LL4 because there are years where there is catch but no LFs but we’ve set this fishery to direct removals.
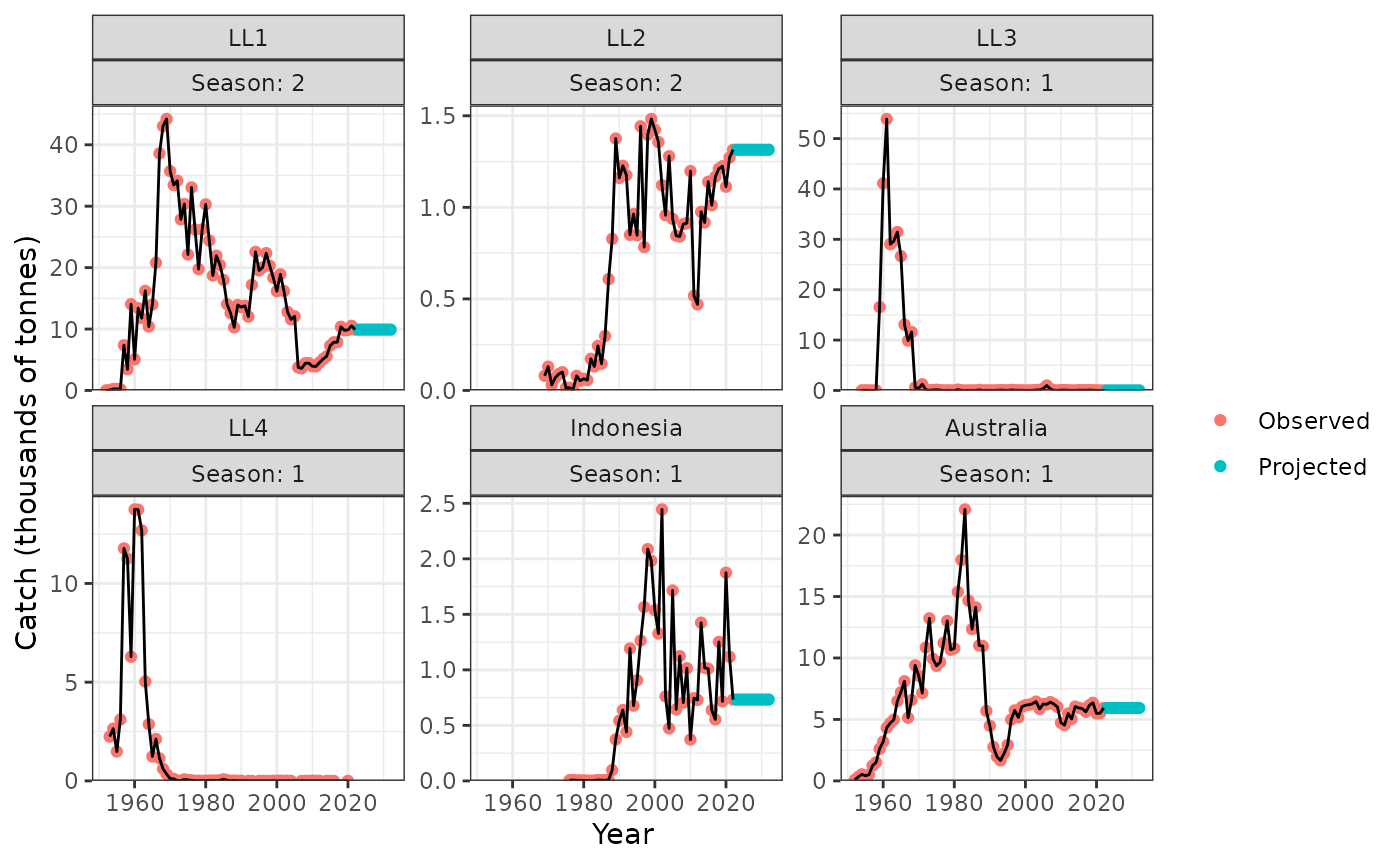
plot_catch(data = data, object = obj)
#> [1] "The maximum catch difference was: 18.98"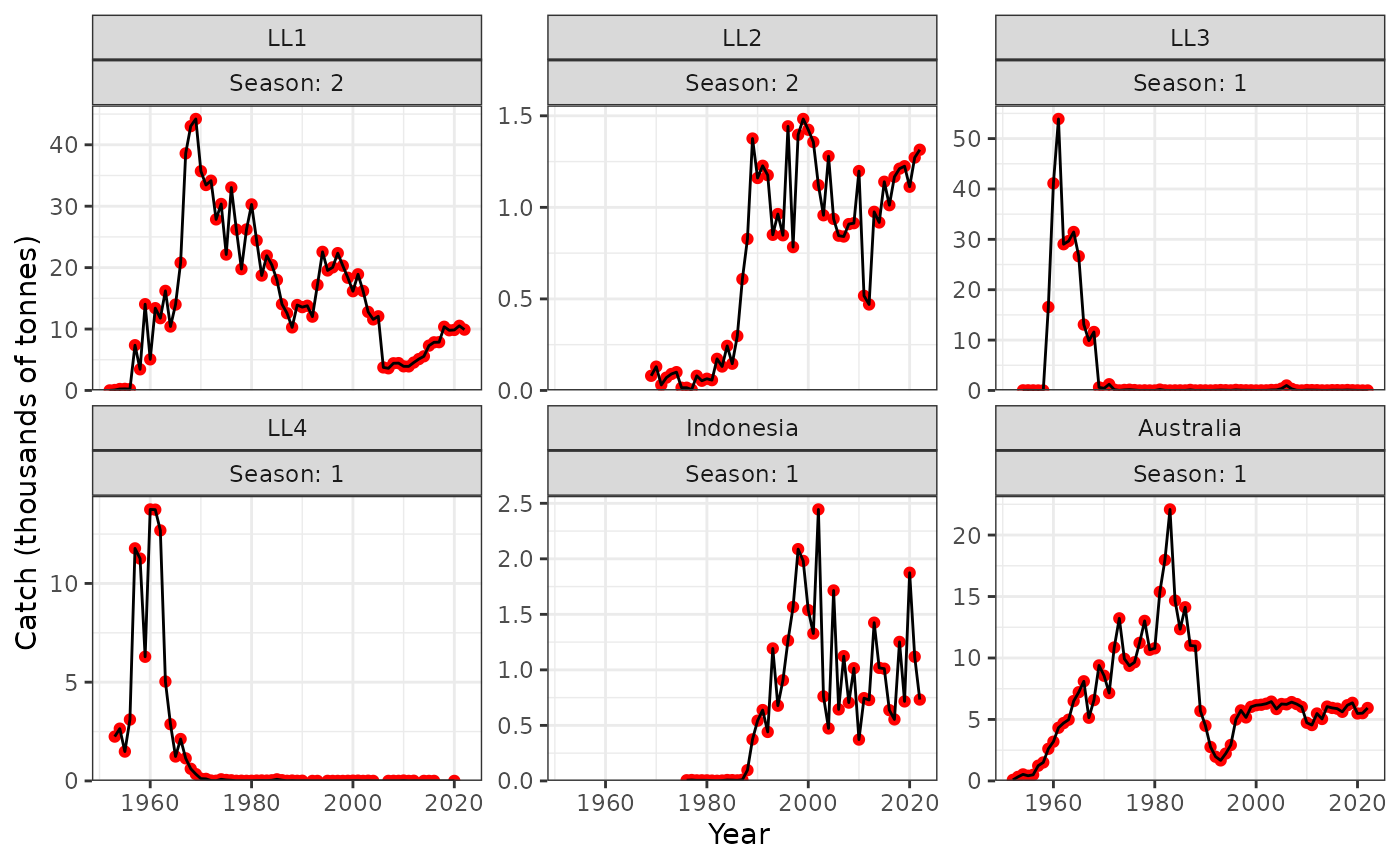
plot_catch(data = data, object = obj, plot_resid = TRUE)
#> [1] "The maximum catch difference was: 18.98"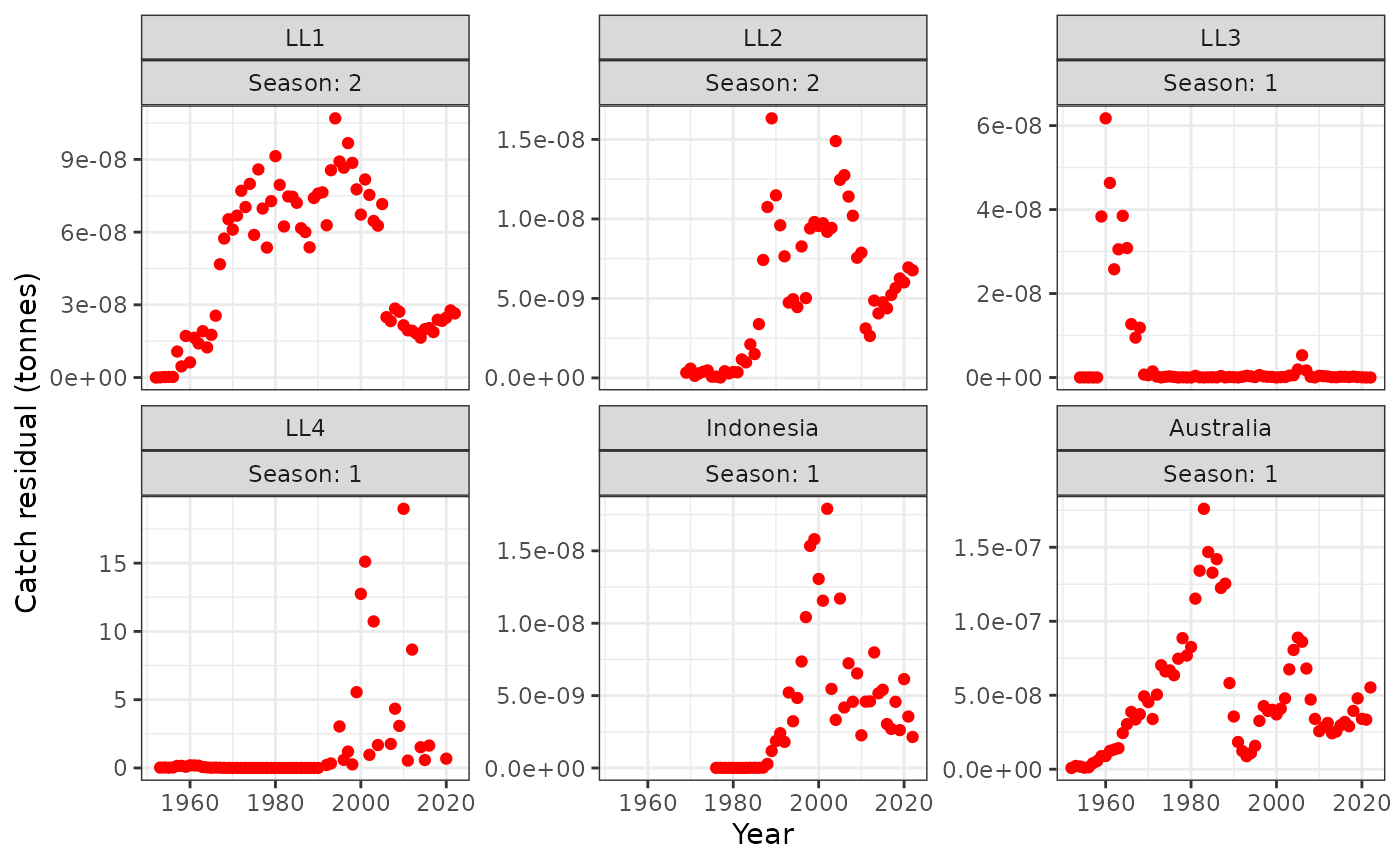
One step ahead (OSA) residuals
One step ahead (OSA) residuals are a replacement for Pearson residuals (Figure~(fig:osa1)).
osa_cpue <- oneStepPredict(obj = obj, observation.name = "cpue_log_obs",
method = "oneStepGeneric", trace = FALSE)
qqnorm(osa_cpue$res); abline(0, 1)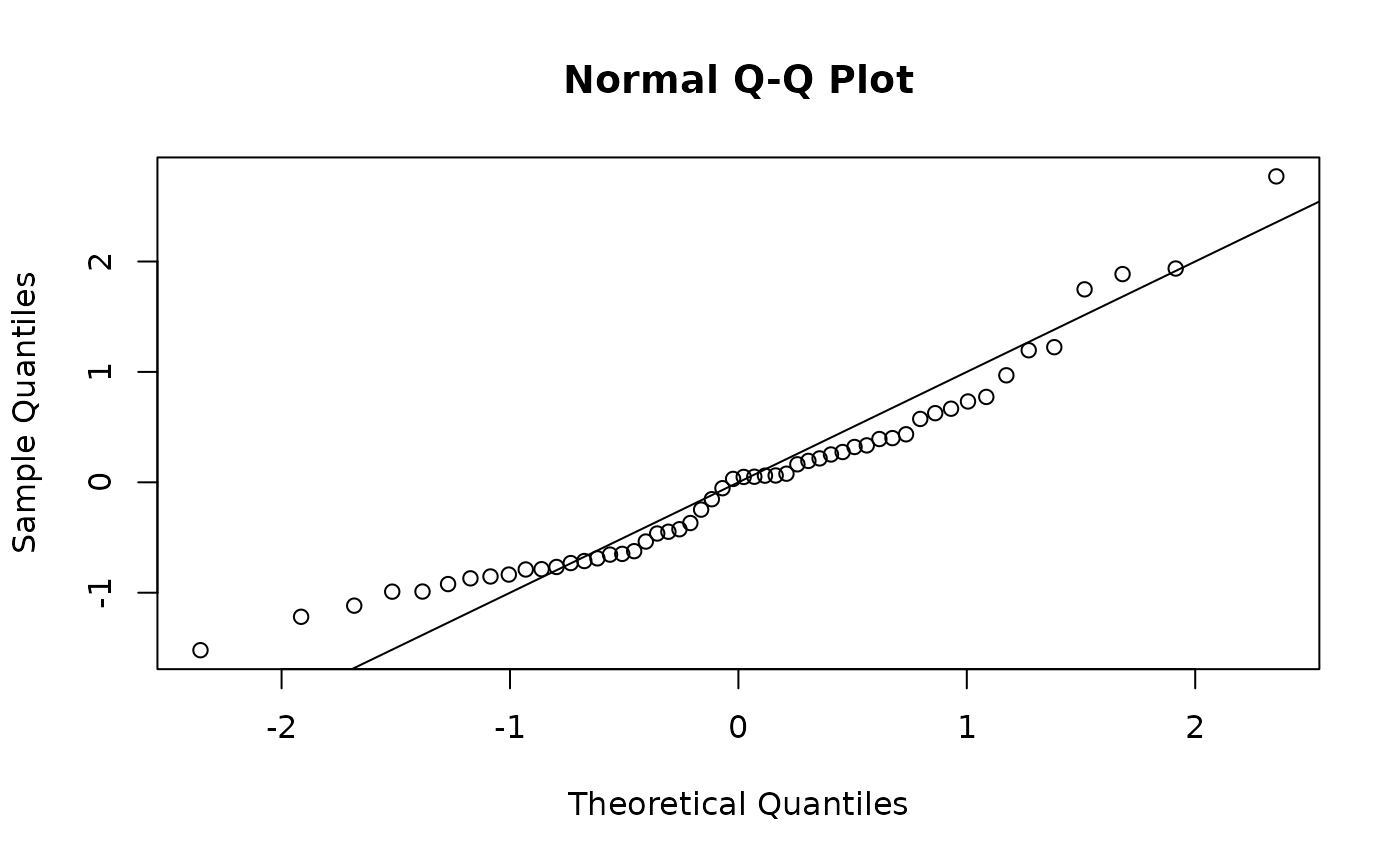
OSA residuals.
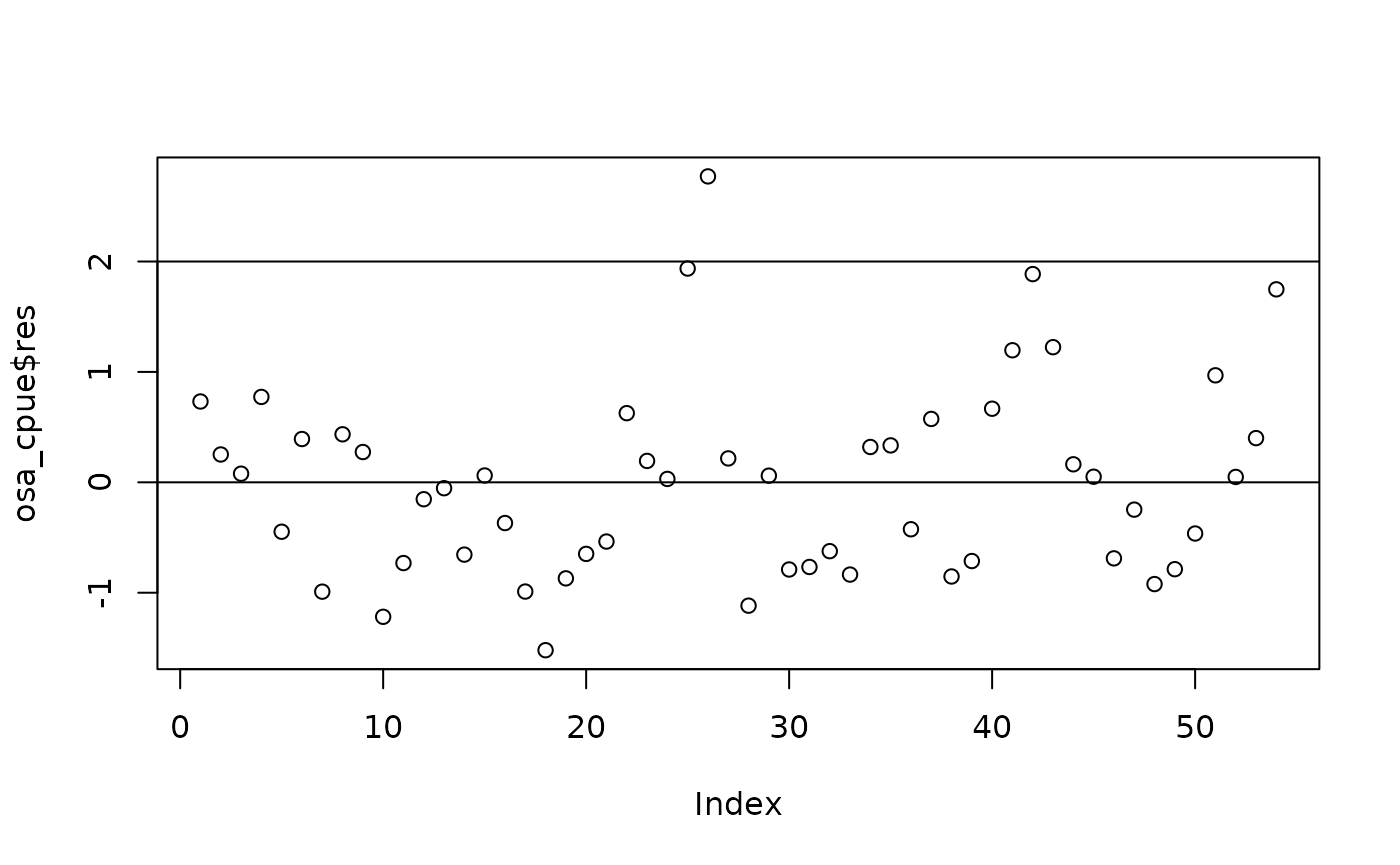
OSA residuals.
# osa <- oneStepPredict(obj = obj, method = "fullGaussian", discrete = FALSE, trace = FALSE)
# osa_troll <- oneStepPredict(obj = obj, observation.name = "troll_log_obs", method = "oneStepGeneric", trace = FALSE)
# qqnorm(osa_troll$res); abline(0, 1)
# plot(osa_troll$res); abline(0, 0)
# osa_aerial <- oneStepPredict(obj = obj, observation.name = "aerial_log_obs", method = "oneStepGeneric", trace = FALSE)
# qqnorm(osa$res); abline(0, 1)
# osa_gt <- oneStepPredict(obj = obj, observation.name = "gt_nrec", method = "oneStepGeneric", discrete = TRUE, trace = FALSE)
# qqnorm(osa_gt$res); abline(0, 1)
# plot(osa_gt$res); abline(-2, 0); abline(0, 0); abline(2, 0)
# osa_hsp <- oneStepPredict(obj = obj, observation.name = "hsp_nK", method = "oneStepGeneric", discrete = TRUE, trace = FALSE)
# qqnorm(osa_hsp$res); abline(0, 1)
# plot(osa_hsp$res); abline(0, 0)
# osa_pop <- oneStepPredict(obj = obj, observation.name = "pop_nP", method = "oneStepGeneric", discrete = TRUE, trace = FALSE)
# qqnorm(osa_pop$res); abline(0, 1)
# plot(osa_pop$res); abline(0, 0)Derived quantities
plot_rec_devs(data = data, object = obj)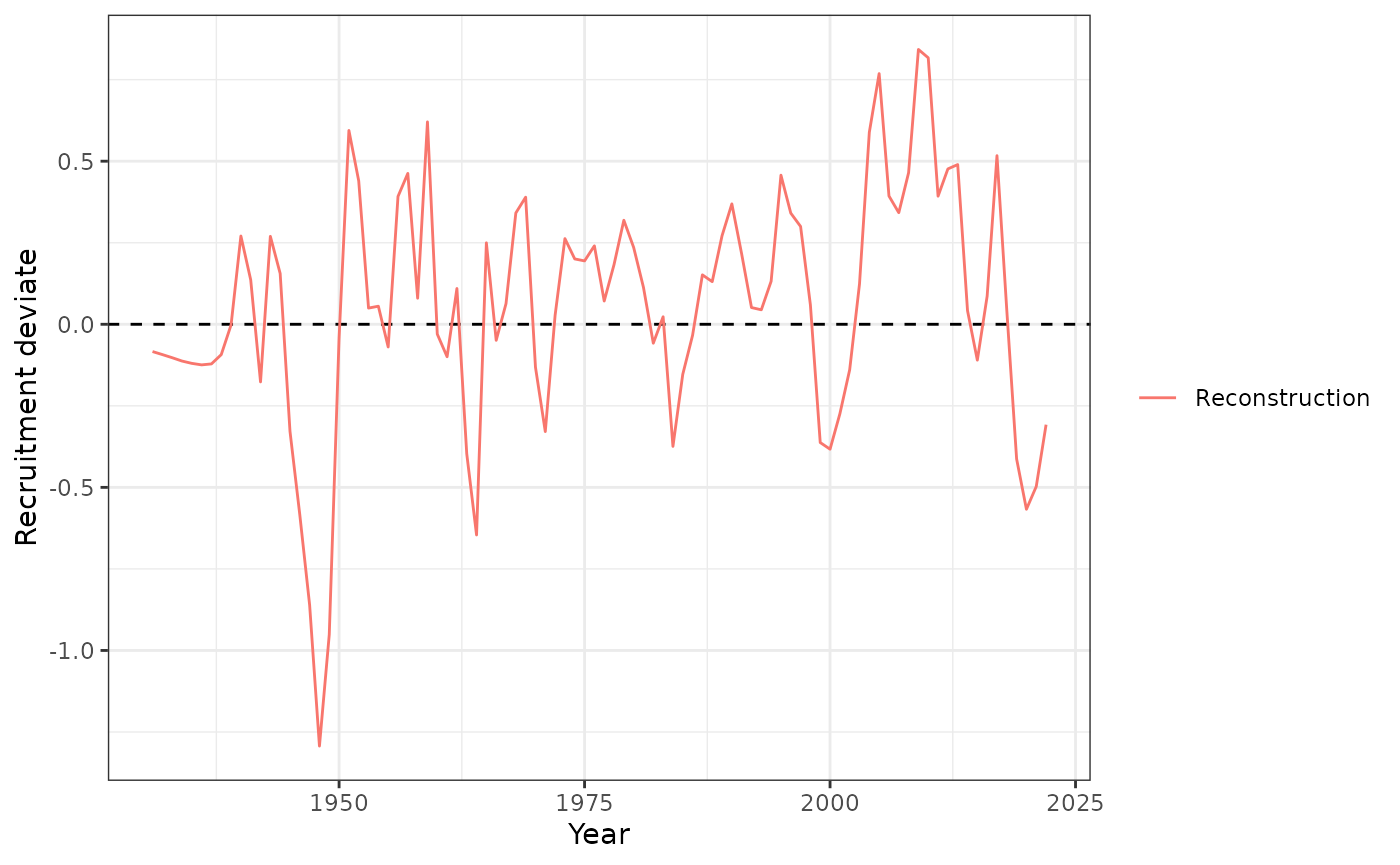
plot_recruitment(data = data, object = obj)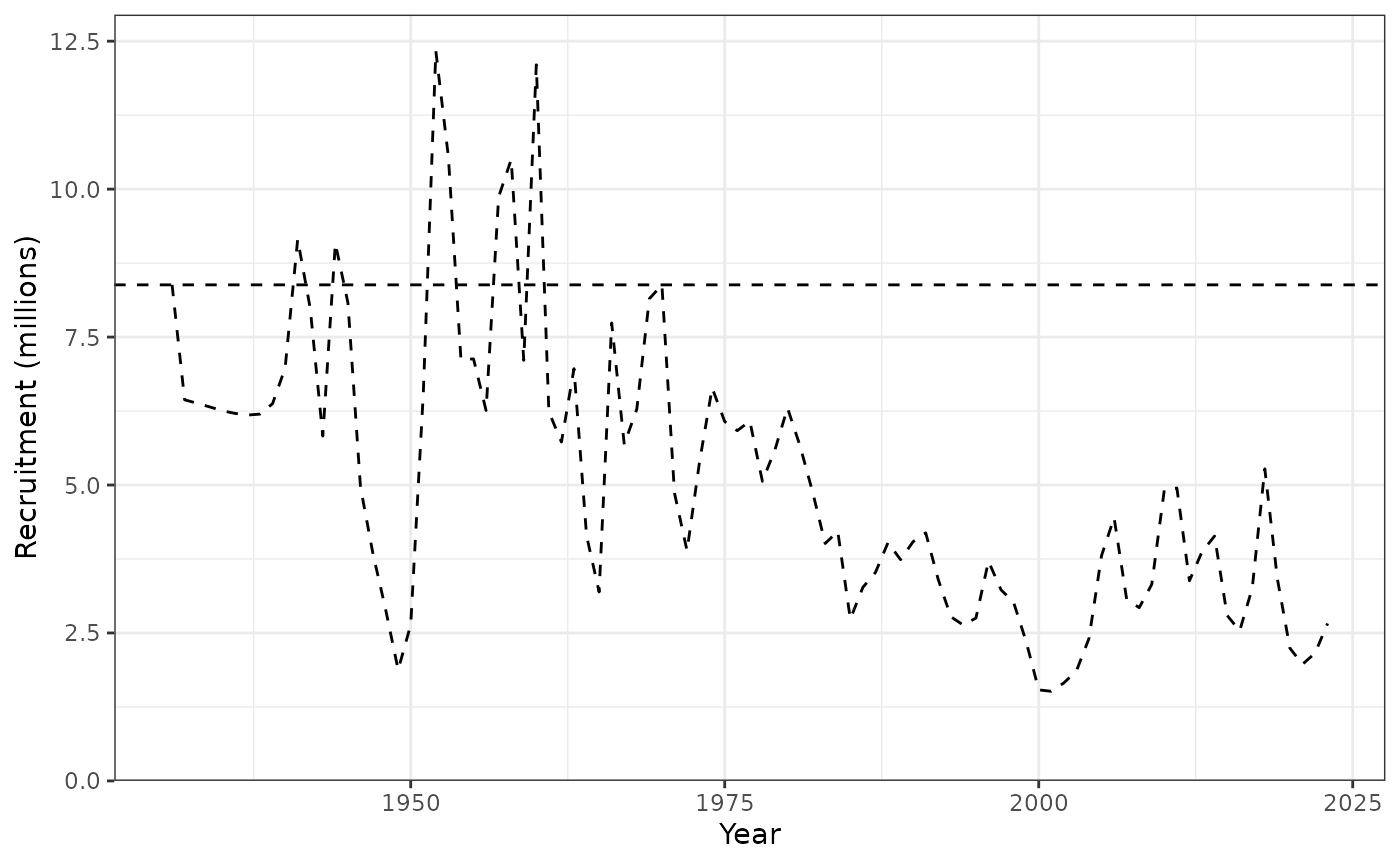
plot_natural_mortality(data = data, object = obj)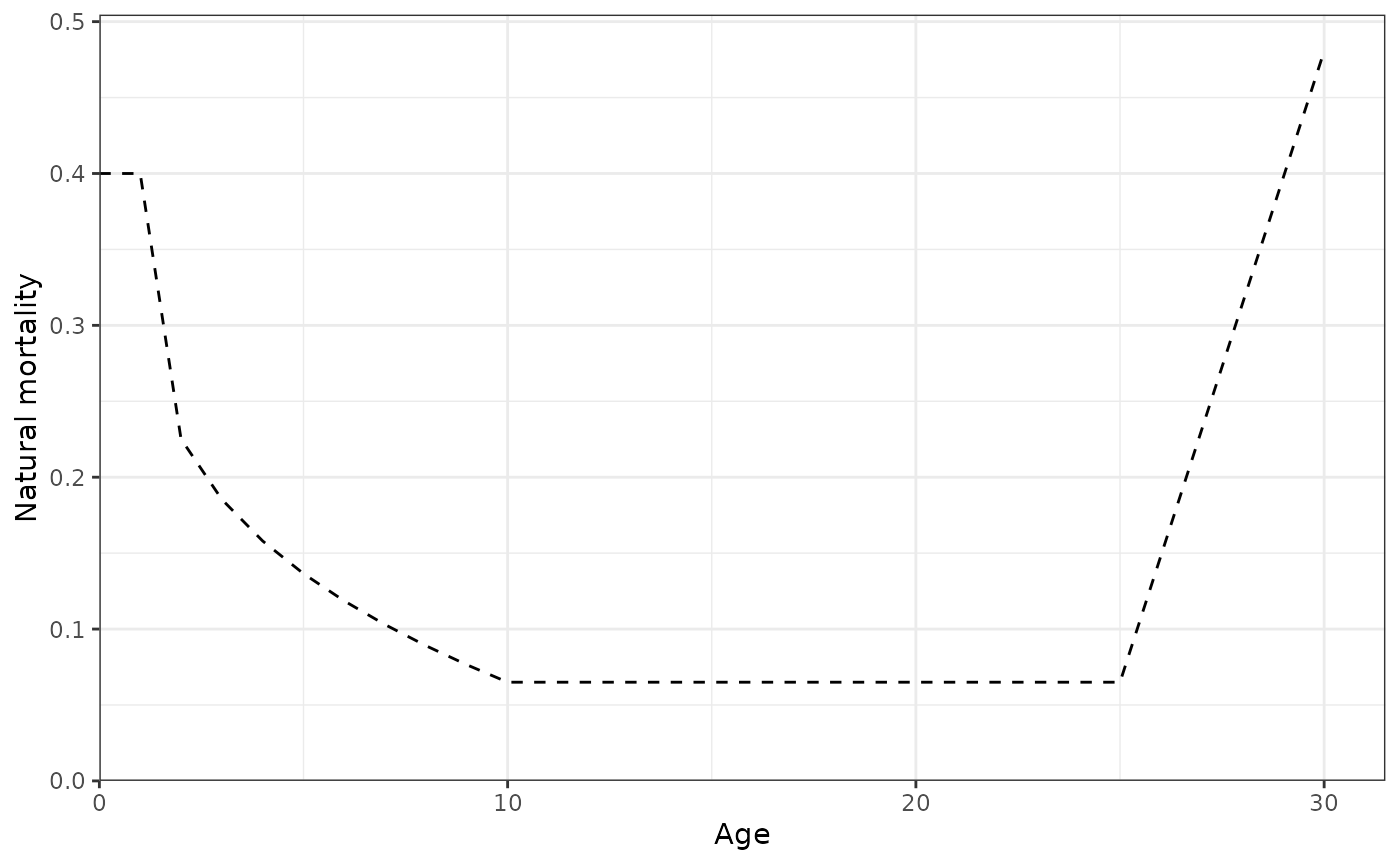
plot_initial_numbers(data = data, object = obj)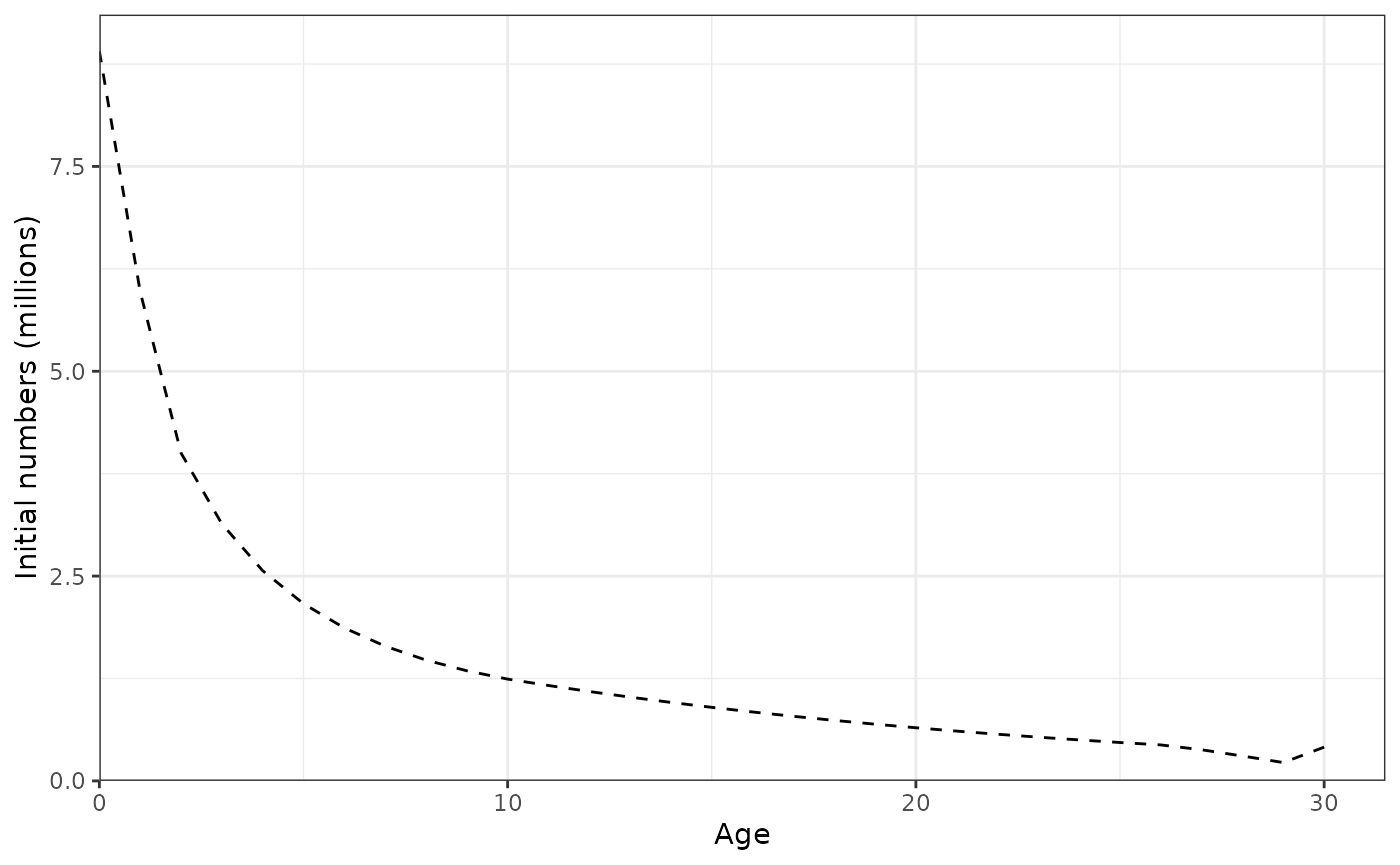
plot_hrate(data = data, object = obj, years = 1990:2010)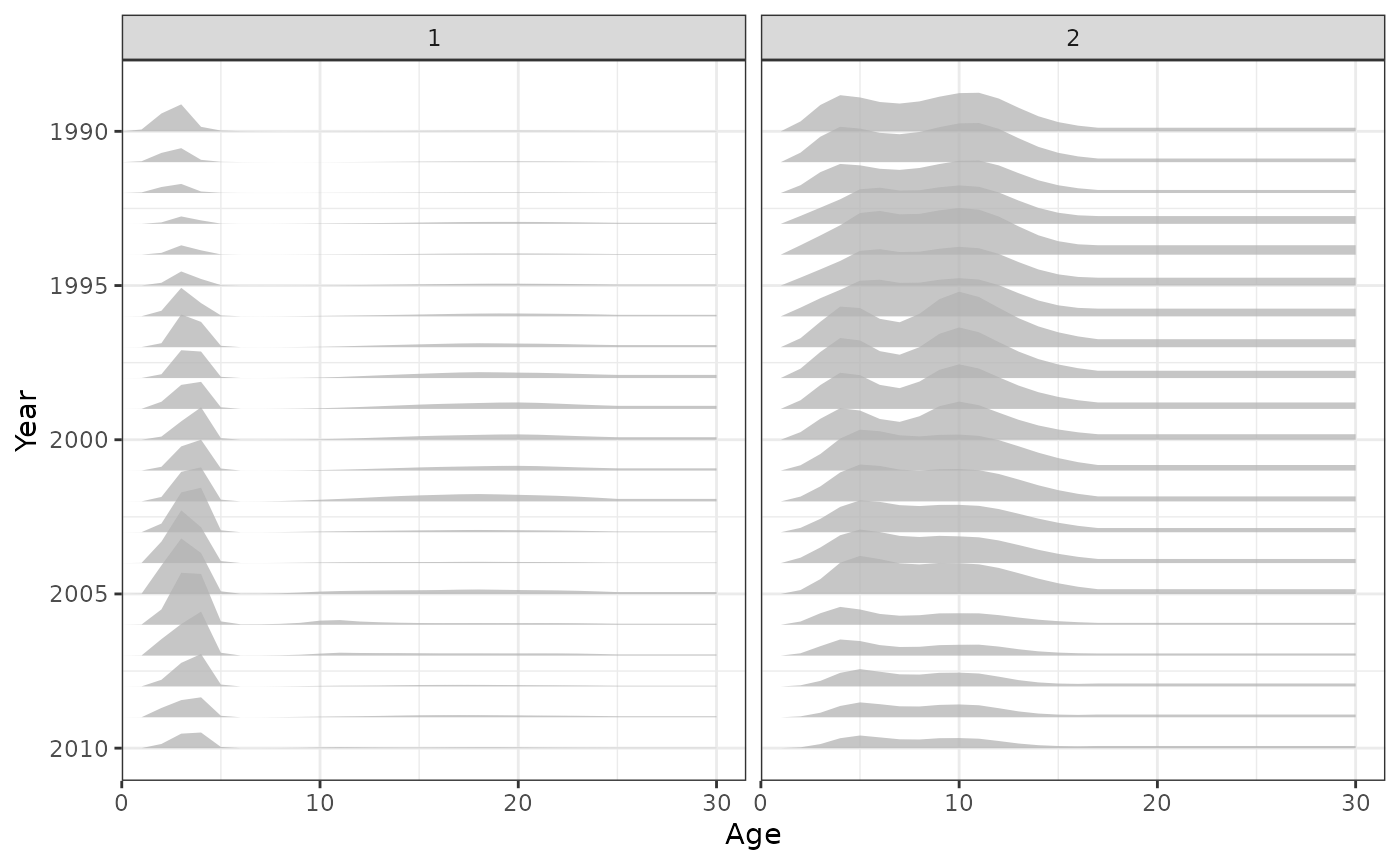
In the plots below, the different coloured blocks of years indicate blocks of parameters, vertical dashed lines indicate the minimum and maximum ages over which selectivity is estimated, and crosses to the left indicate that LF data is available during that year.
# yrs <- c(1952, 1953, 1954, seq(1955, 2020, 5), 2021, 2022)
yrs <- c(1952:2022)
plot_selectivity(data = data, object = obj, years = yrs, fisheries = "LL1")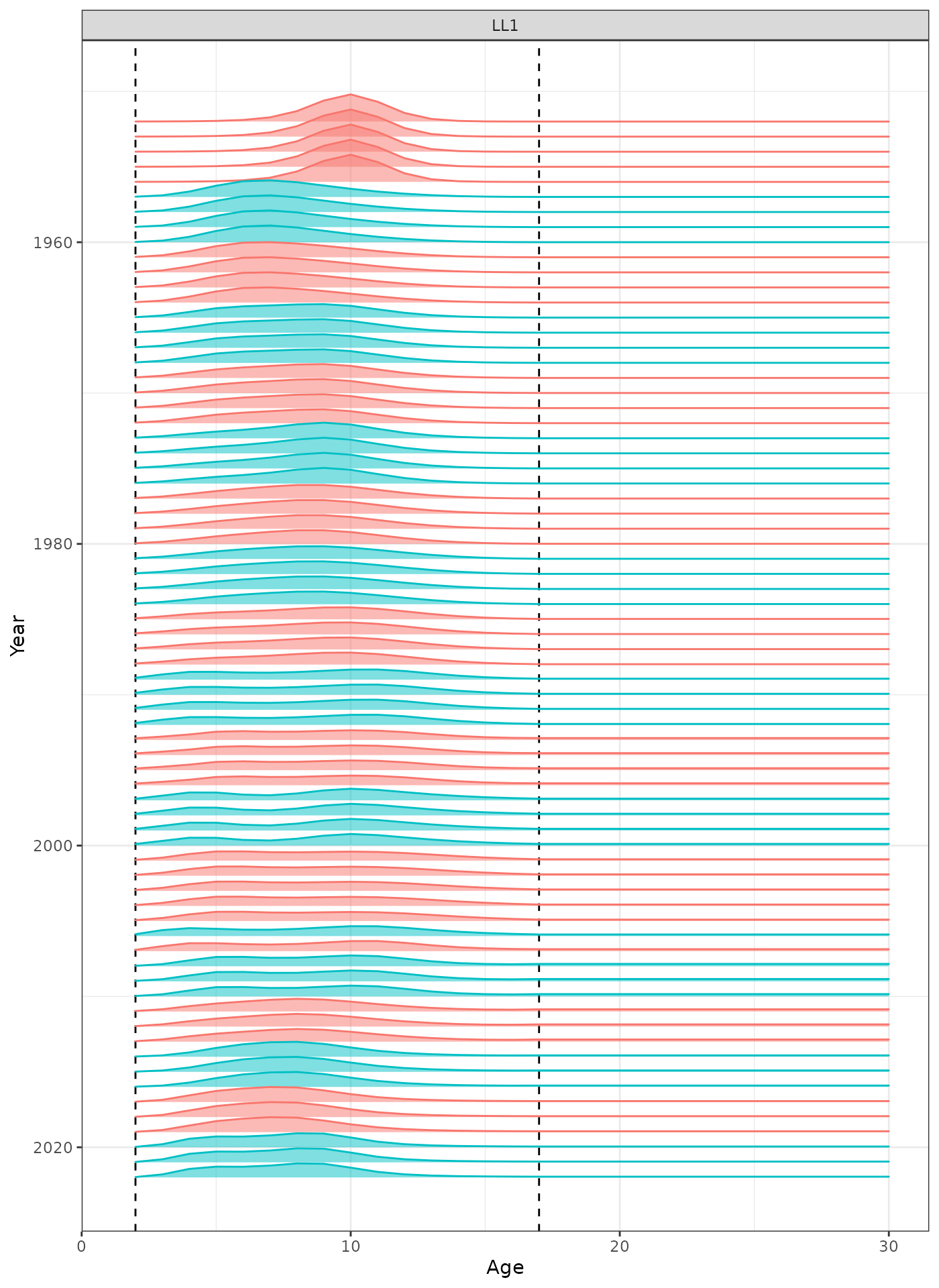
Selectivity at age by year for the LL1 fleet.
plot_selectivity(data = data, object = obj, years = yrs, fisheries = "LL2")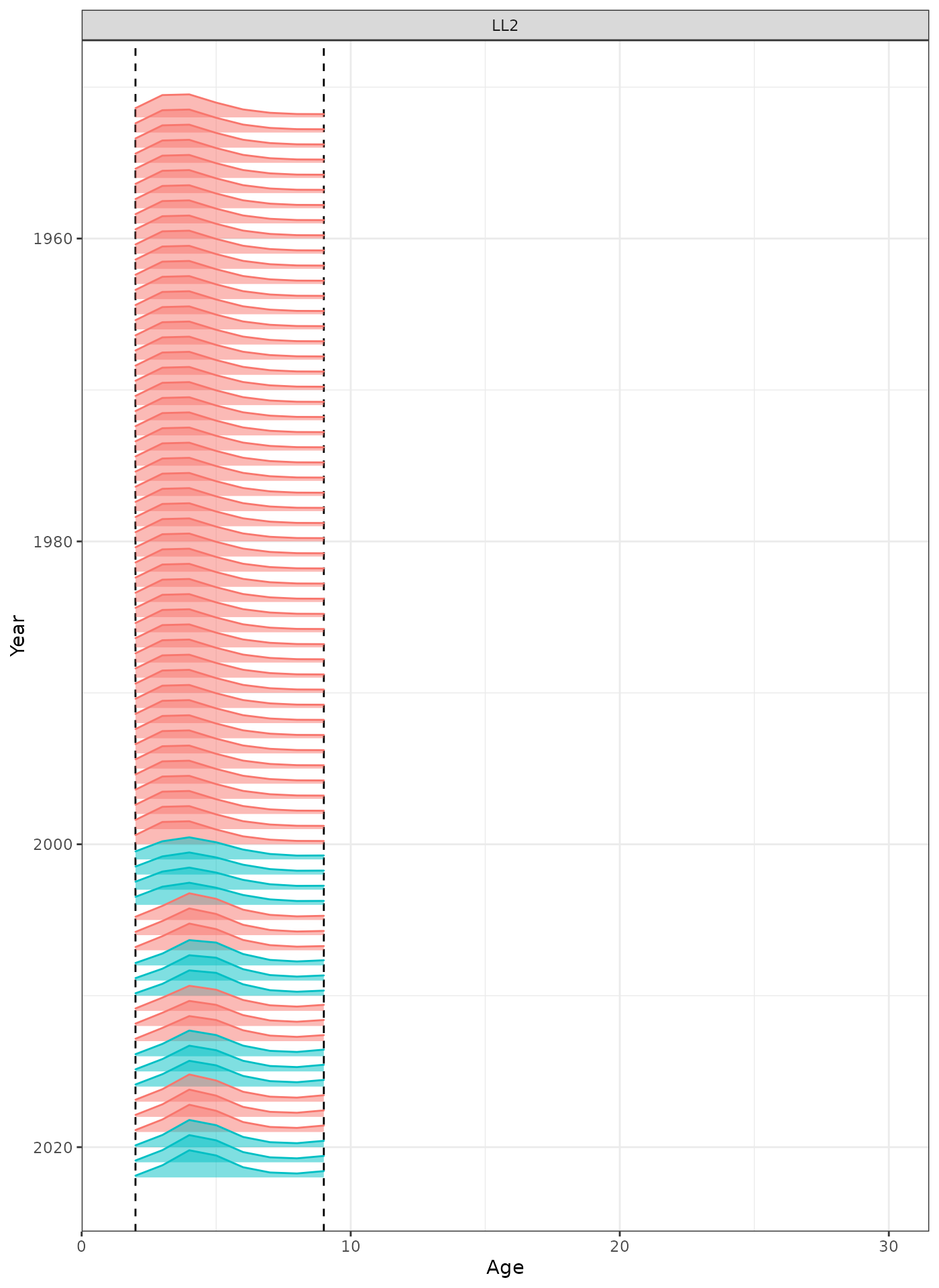
Selectivity at age by year for the LL2 fleet.
plot_selectivity(data = data, object = obj, years = yrs, fisheries = "LL3")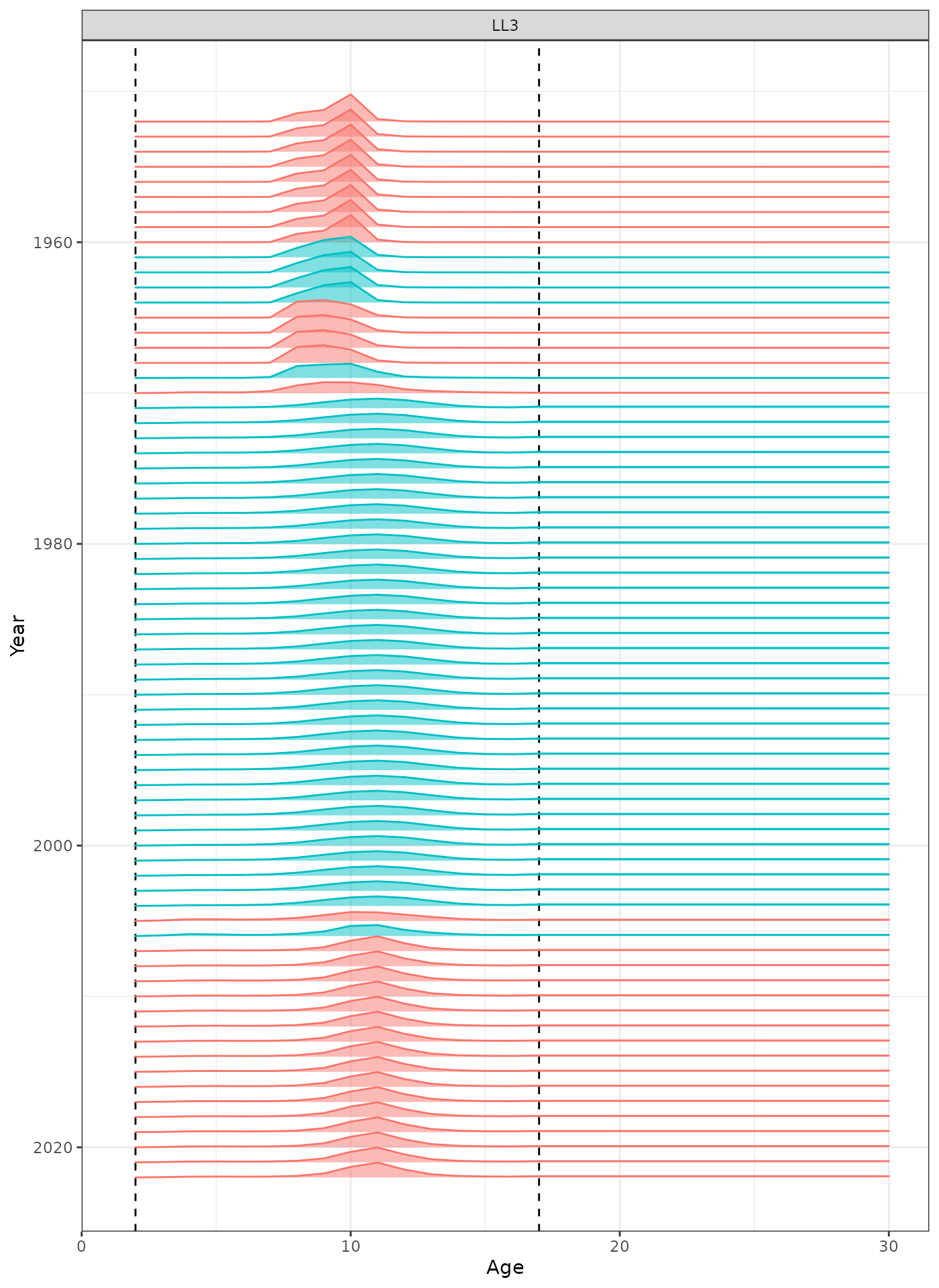
Selectivity at age by year for the LL3 fleet.
# plot_selectivity(data = data, object = obj, years = yrs, fisheries = "LL4")
plot_selectivity(data = data, object = obj, years = yrs, fisheries = "Indonesian")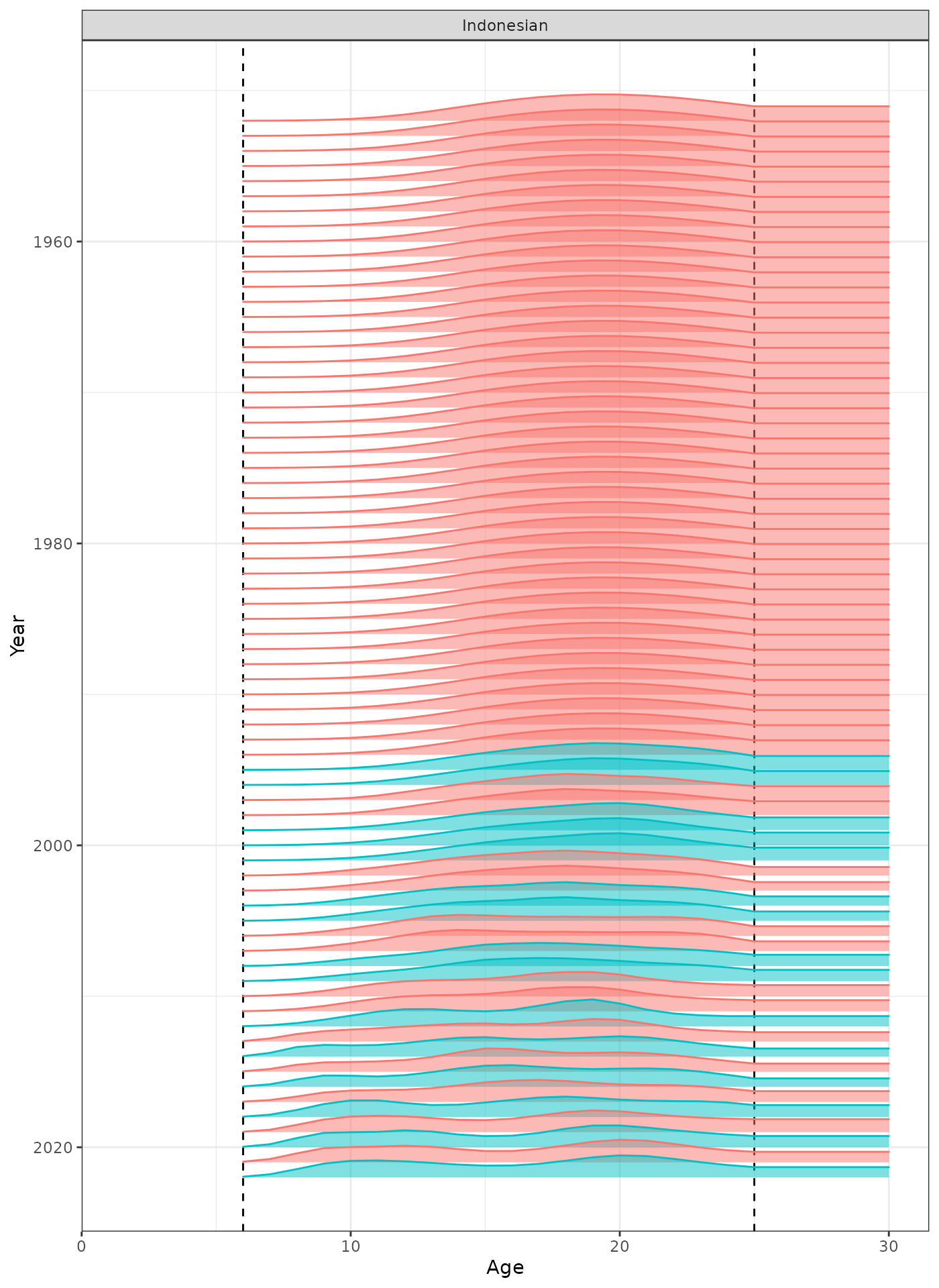
Selectivity at age by year for the Indonesian fleet.
plot_selectivity(data = data, object = obj, years = yrs, fisheries = "Australian")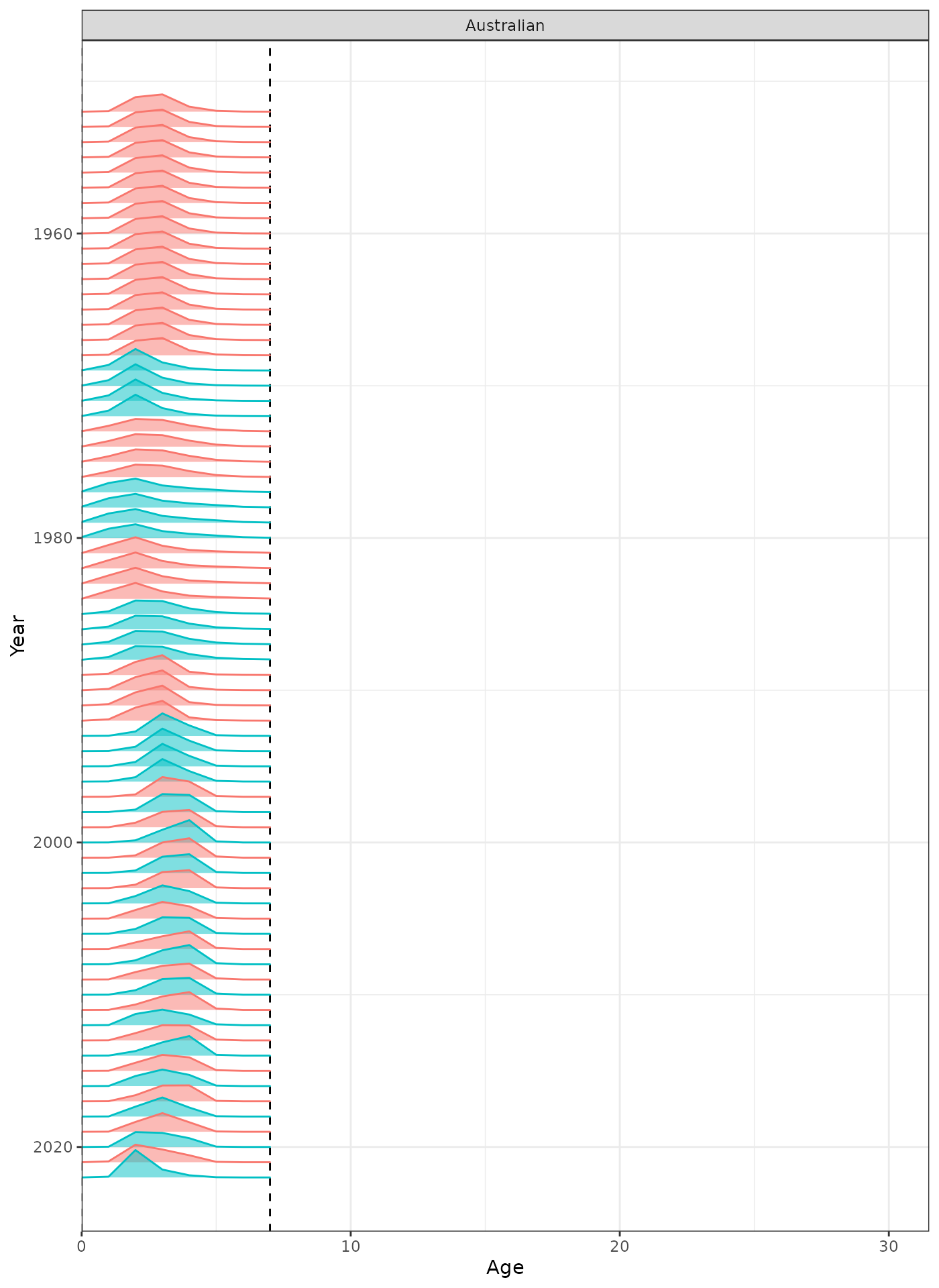
Selectivity at age by year for the Australian fleet.
plot_selectivity(data = data, object = obj, years = yrs, fisheries = "CPUE")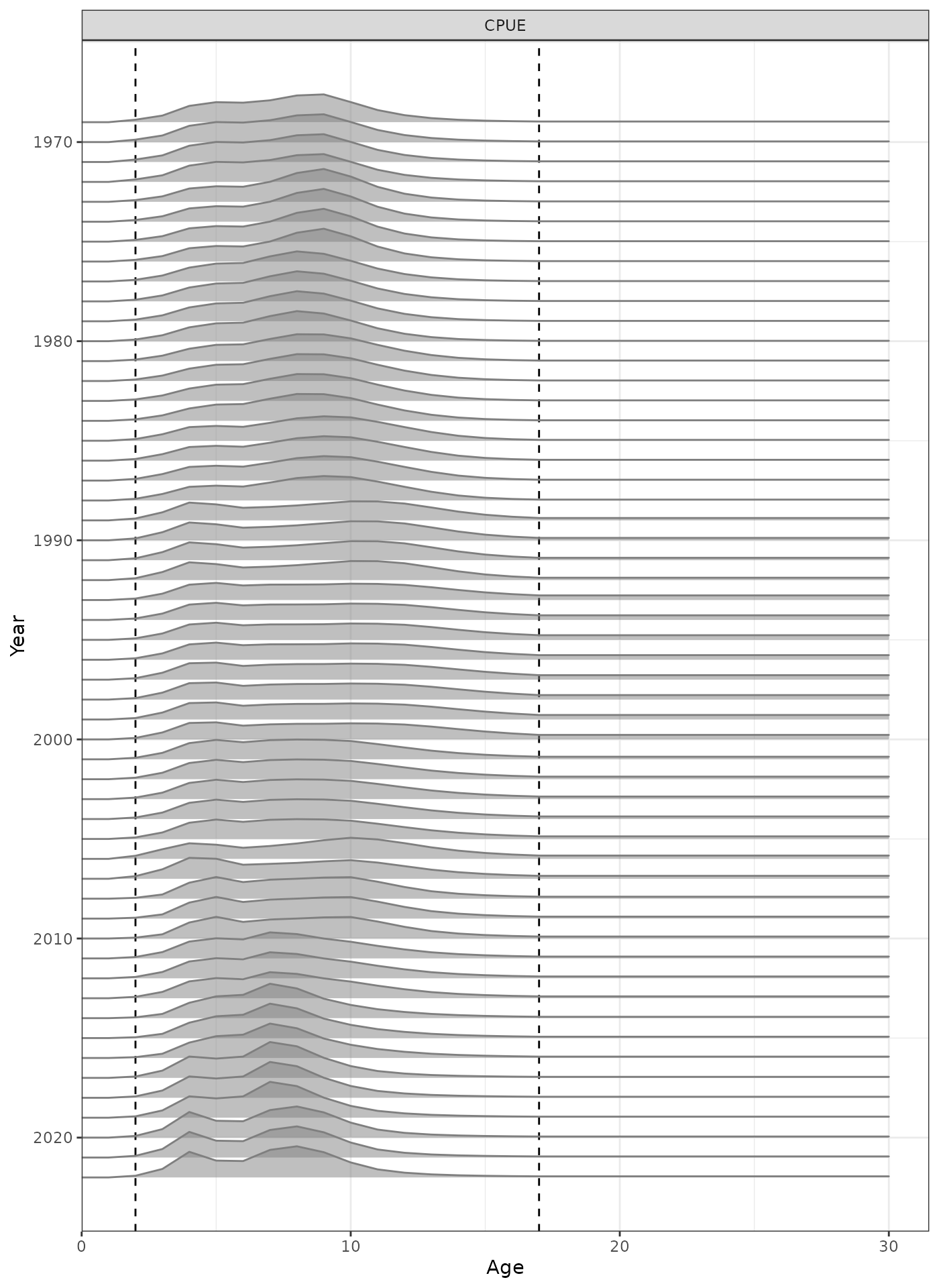
Selectivity at age by year for the CPUE index.
plot_biomass_spawning(data = data, object = obj)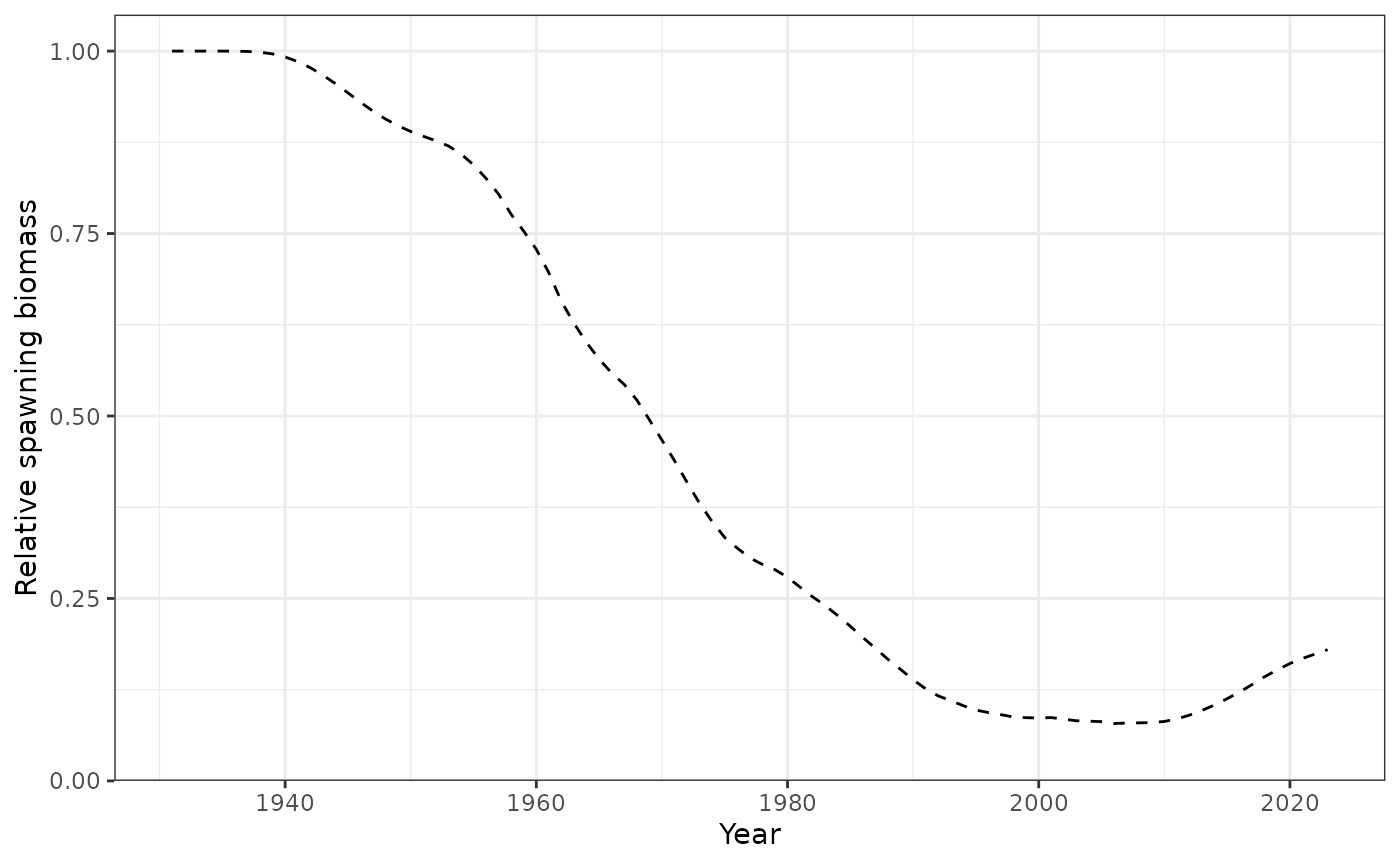
Spawning biomass by year.
Likelihood profiles
Likelihood profiles can be done for any model parameter using the
function sbtprofile. The ytol argument defines
the range of likelihood values to explore. A profile can be run using
either using name or lincomb arguments, where
the latter can be used if there are multiple parameters with the same
name (e.g., par_log_sel_1).
# ytol <- 9
# prof_B0 <- sbtprofile(obj = obj, name = "par_log_B0", ytol = ytol)
# prof_m4 <- sbtprofile(obj = obj, name = "par_log_m4", ytol = ytol)
# prof_m30 <- sbtprofile(obj = obj, name = "par_log_m30", ytol = ytol)
# lincomb <- numeric(length(obj$par)); lincomb[6] <- 1
# prof_sel1 <- sbtprofile(obj = obj, lincomb = lincomb, ytol = ytol)
# save(prof_B0, prof_m4, prof_m30, prof_sel1, file = "profiles.rda")
load("profiles.rda")Note that the profile for B0 did not work very well, likelihood for LFs was zero when B0 was too low. Need to look into this.
head(prof_B0)
obj$env$last.par.best[names(obj$par) == "par_log_B0"]
plot_profile(x = prof_B0 %>% filter(is.finite(value)), xlab = "B0")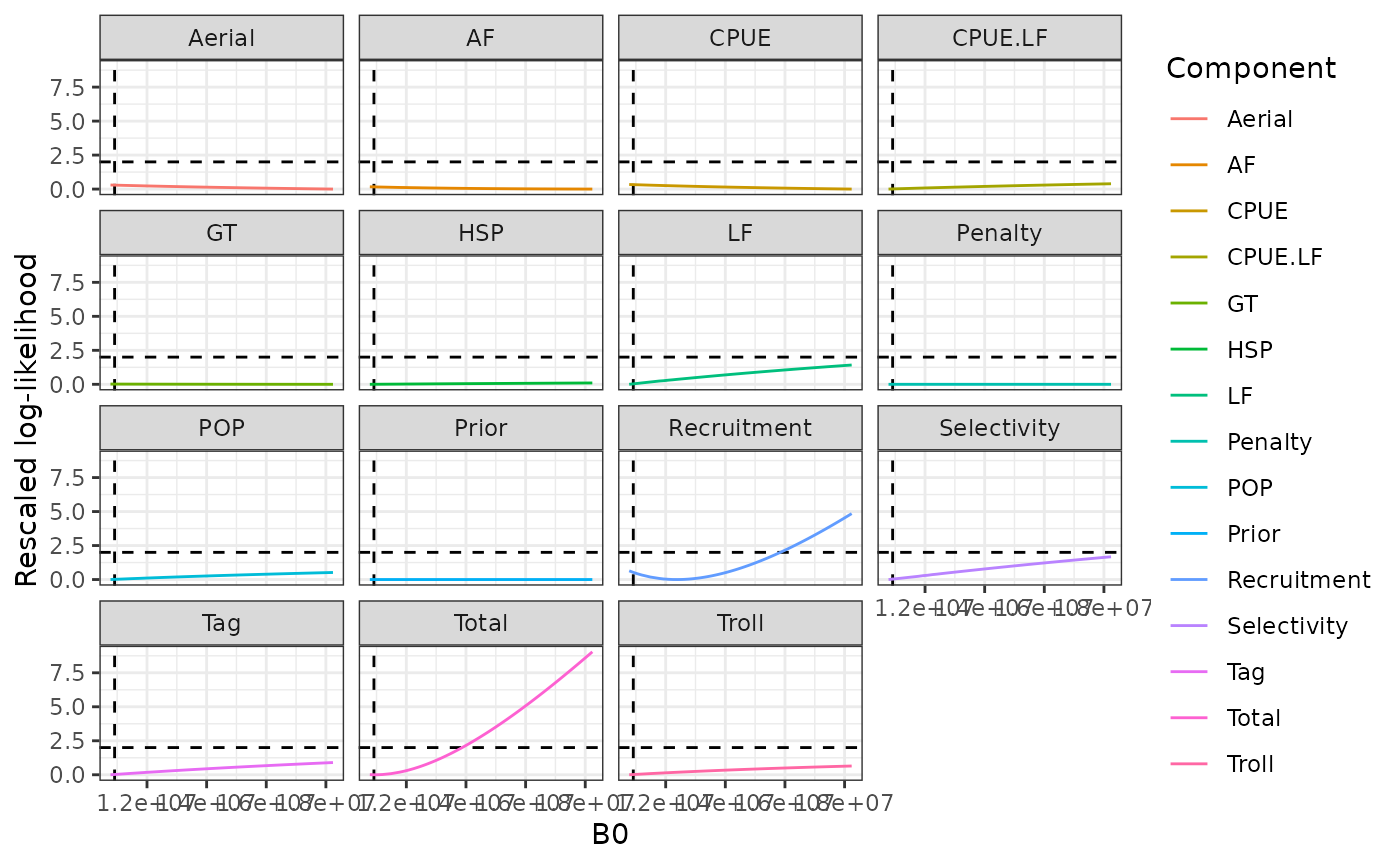
Likelihood profile for B0.
plot_profile(x = prof_m4, xlab = "M4")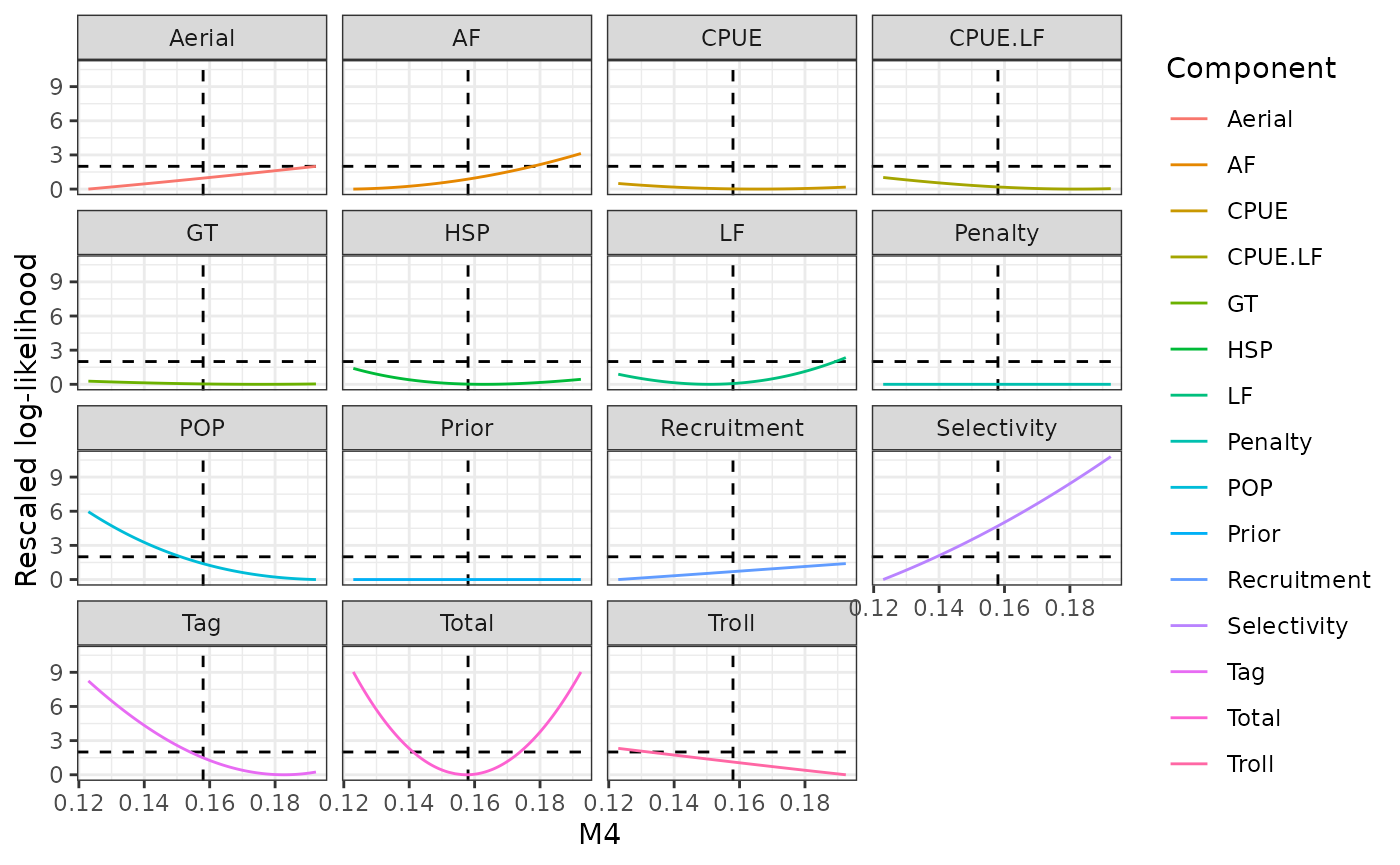
Likelihood profile for M4.
plot_profile(x = prof_m30, xlab = "M30")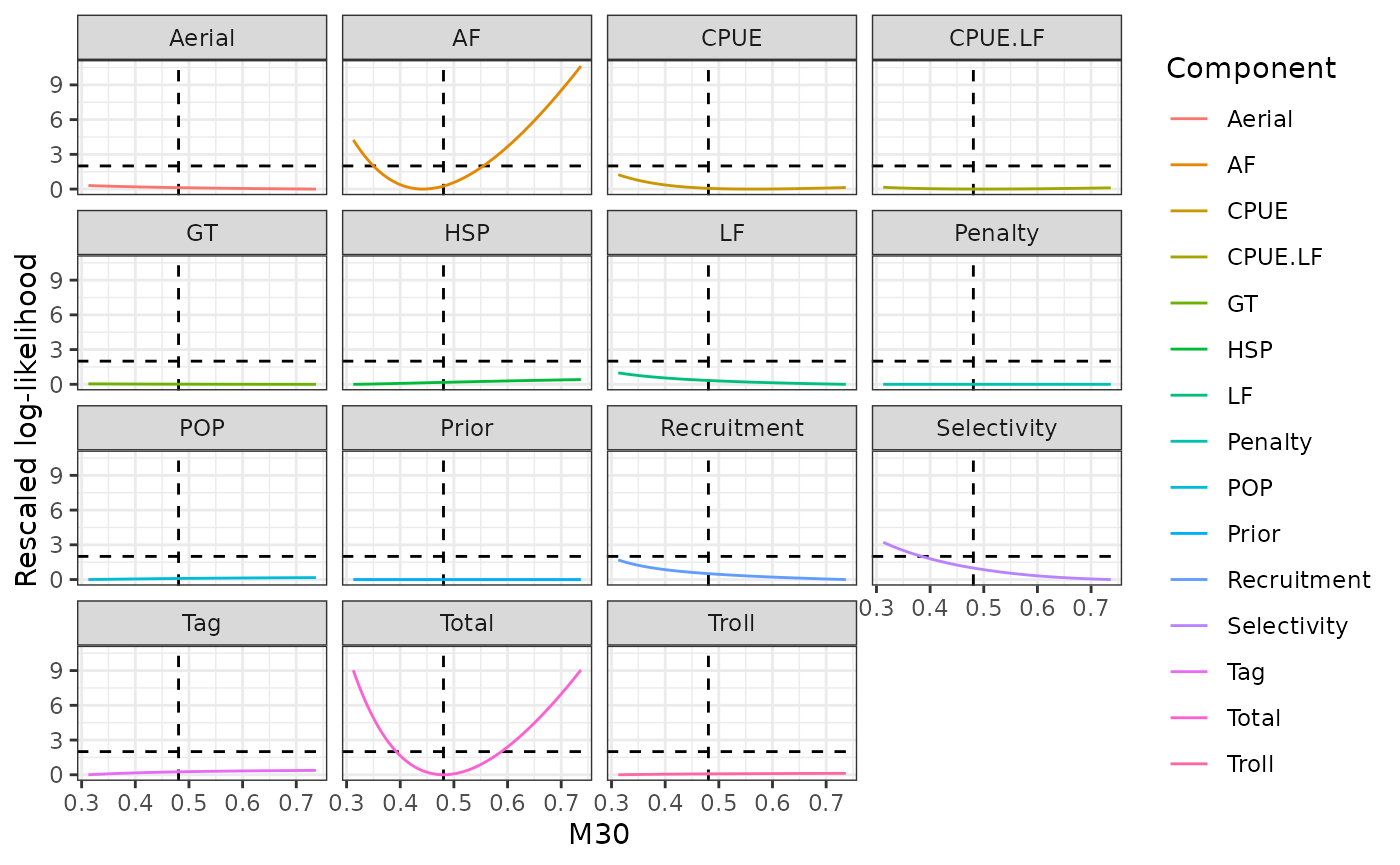
Likelihood profile for M30.
plot_profile(x = prof_sel1, xlab = "Sel1")
#> Warning: Removed 15 rows containing missing values or values outside the scale range
#> (`geom_vline()`).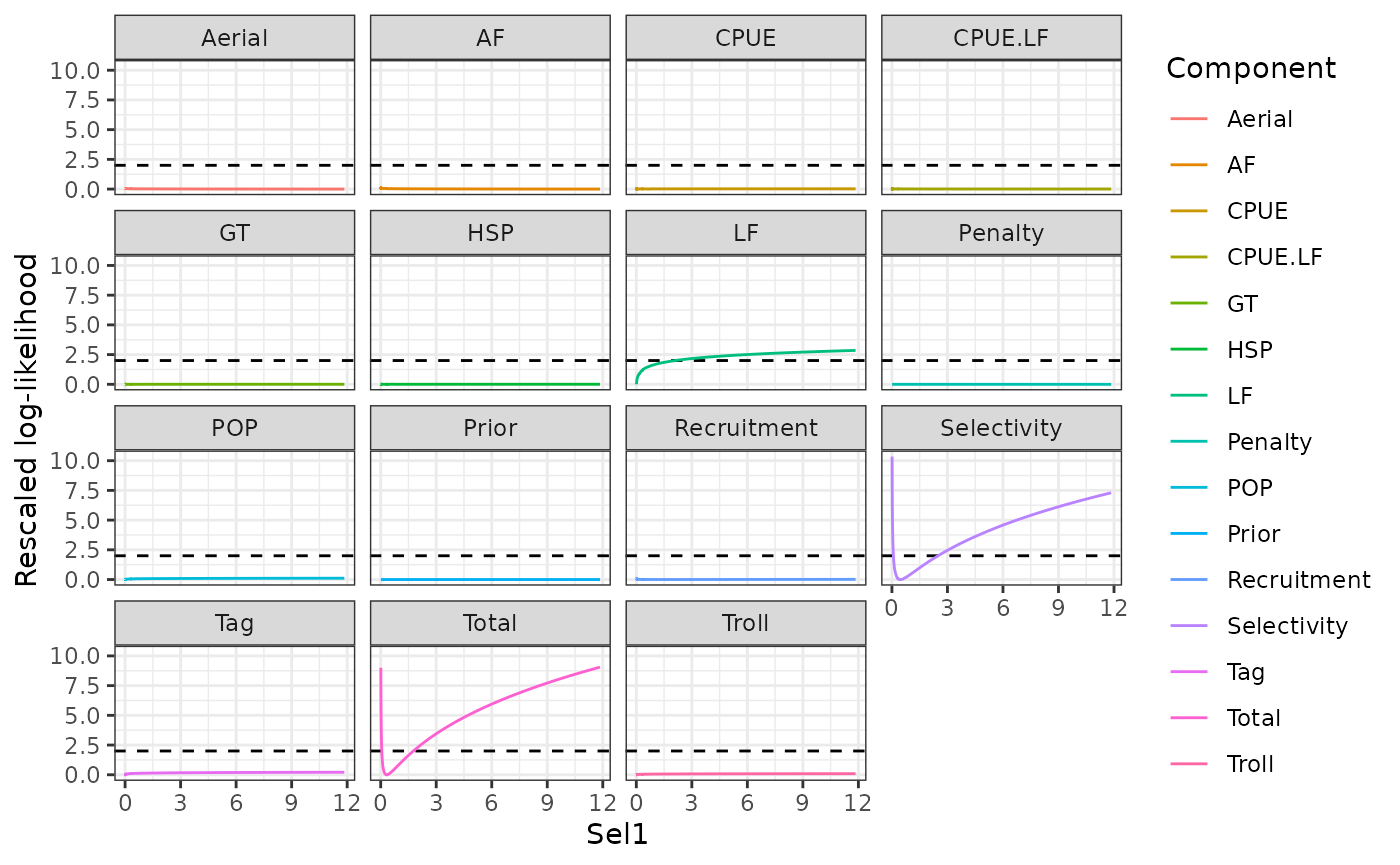
Likelihood profile for one of the selectivity parameters.
Bayesian inference
Bayesian inference can be done using the adnuts
package.
library(adnuts)
#> Loading required package: StanEstimators
# mcmc <- sample_snuts(
# obj = obj, metric = "auto", init = "last.par.best",
# # lower = bnd$lower, upper = bnd$upper, # these bounds dont seem to work
# # skip_optimization = TRUE, # Can skip for Jacobians
# num_samples = 1000, num_warmup = 750, chains = 4, cores = 4,
# control = list(adapt_delta = 0.99), globals = sbt_globals()
# )
# save(mcmc, file = "mcmc.rda")
load("mcmc.rda")
plot_sampler_params(fit = mcmc, plot = TRUE)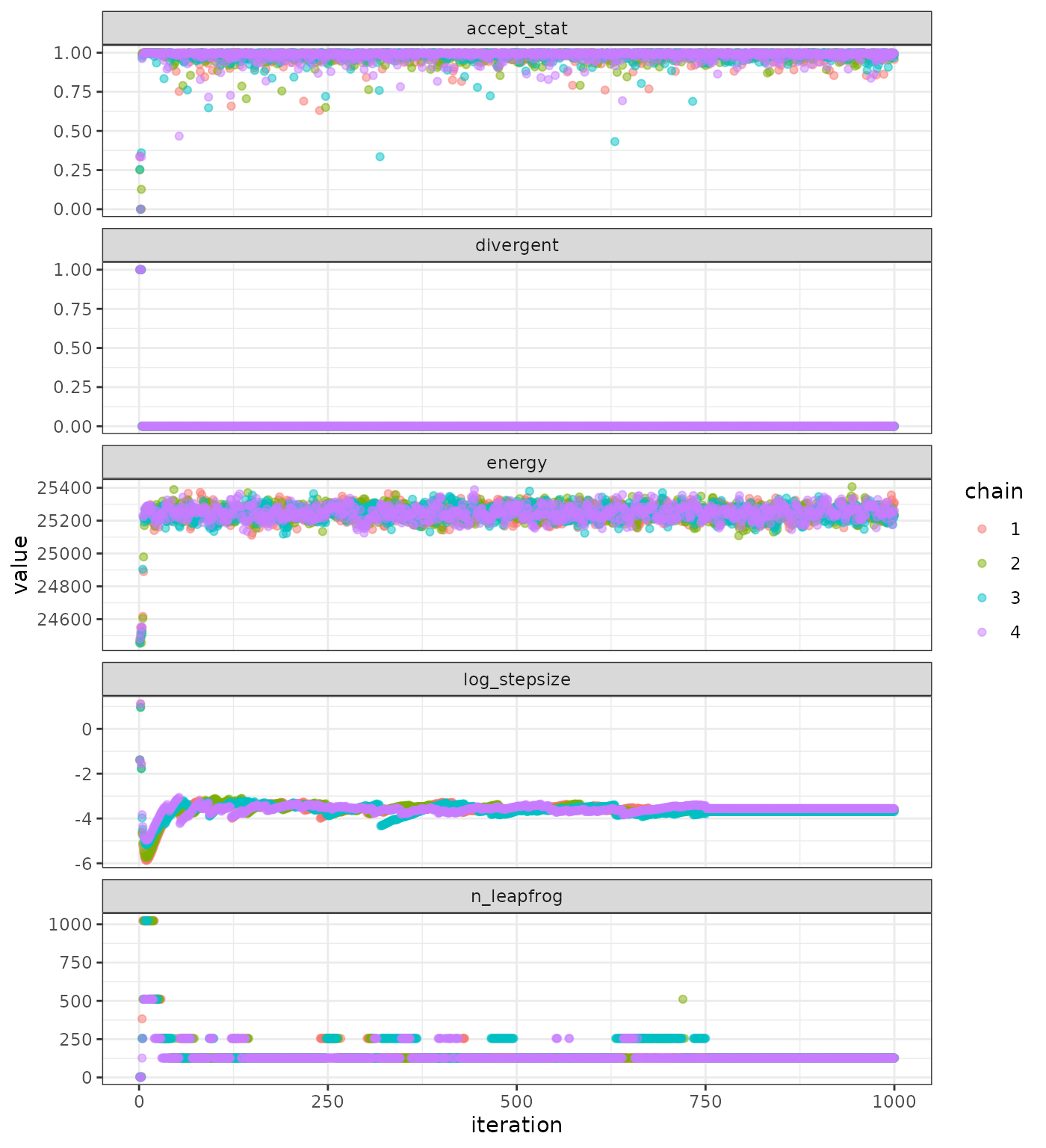
Sampler parameters.
# decamod::pairs_rtmb(fit = mcmc, order = "slow", pars = 1:5)
# decamod::pairs_rtmb(fit = mcmc, order = "mismatch", pars = 1:5)
# decamod::pairs_rtmb(fit = mcmc, order = "fast", pars = 1:5)
plot_uncertainties(fit = mcmc, log = TRUE, plot = TRUE)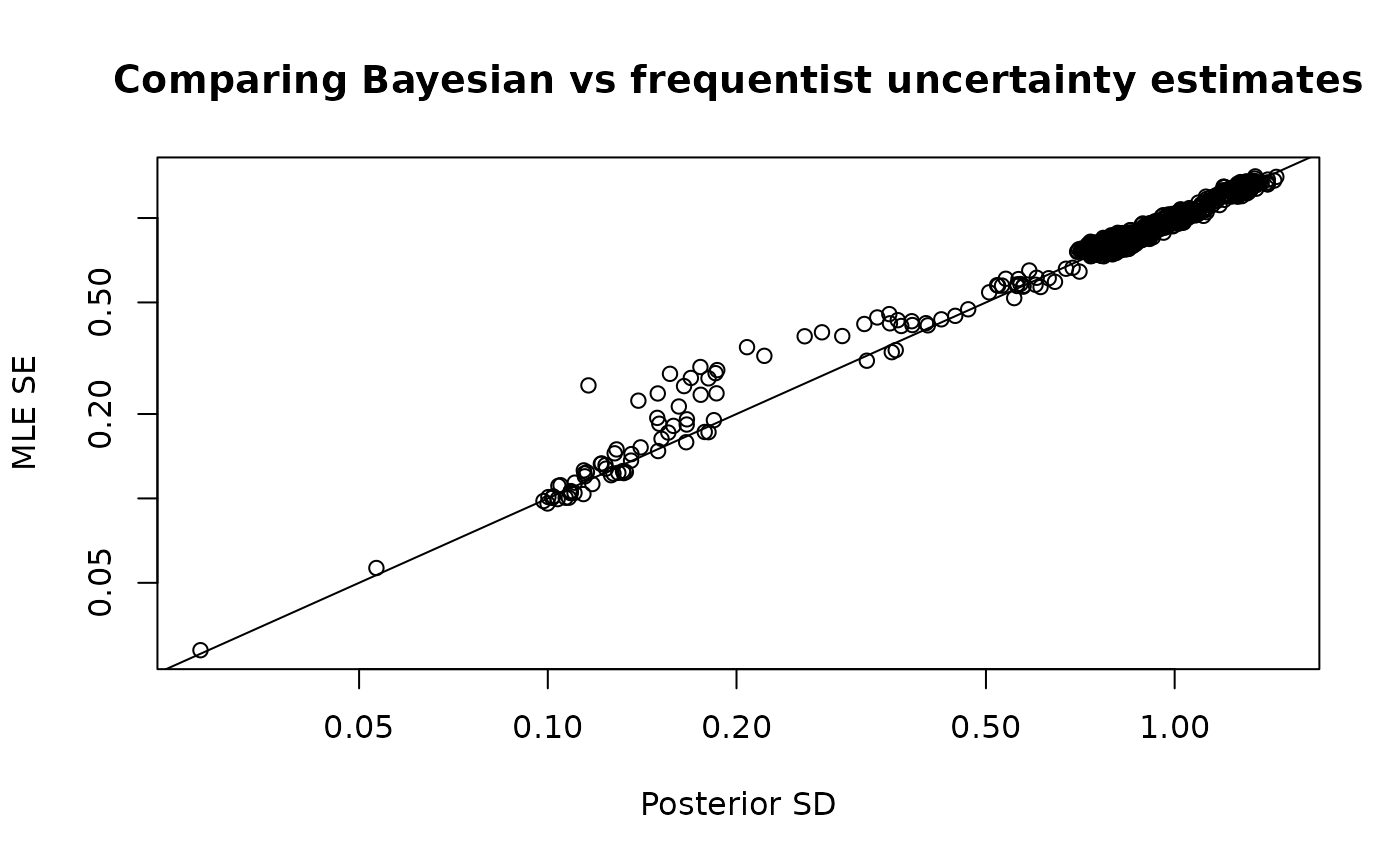
Comparison of Bayesian and frequentist uncertainty estimates.
Projection
Projections can be based on MLE or MCMC.
Recruitment
Recruitment projections can be done the
project_recruitment function. Can be done using using
Autoregressive Integrated Moving Average (ARIMA) models by setting
arima = TRUE.
n_proj <- 10
proj <- project_recruitment(
data = data, obj = obj, mcmc = NULL, first_yr = 1931, last_yr = 2022,
n_proj = n_proj, n_iter = 10, arima = TRUE, max.p = 5)
#> | | | 0% | |======== | 11% | |================ | 22% | |======================= | 33% | |=============================== | 44% | |======================================= | 56% | |=============================================== | 67% | |====================================================== | 78% | |============================================================== | 89% | |======================================================================| 100%
plot_rec_devs(data = data, object = obj, proj = proj)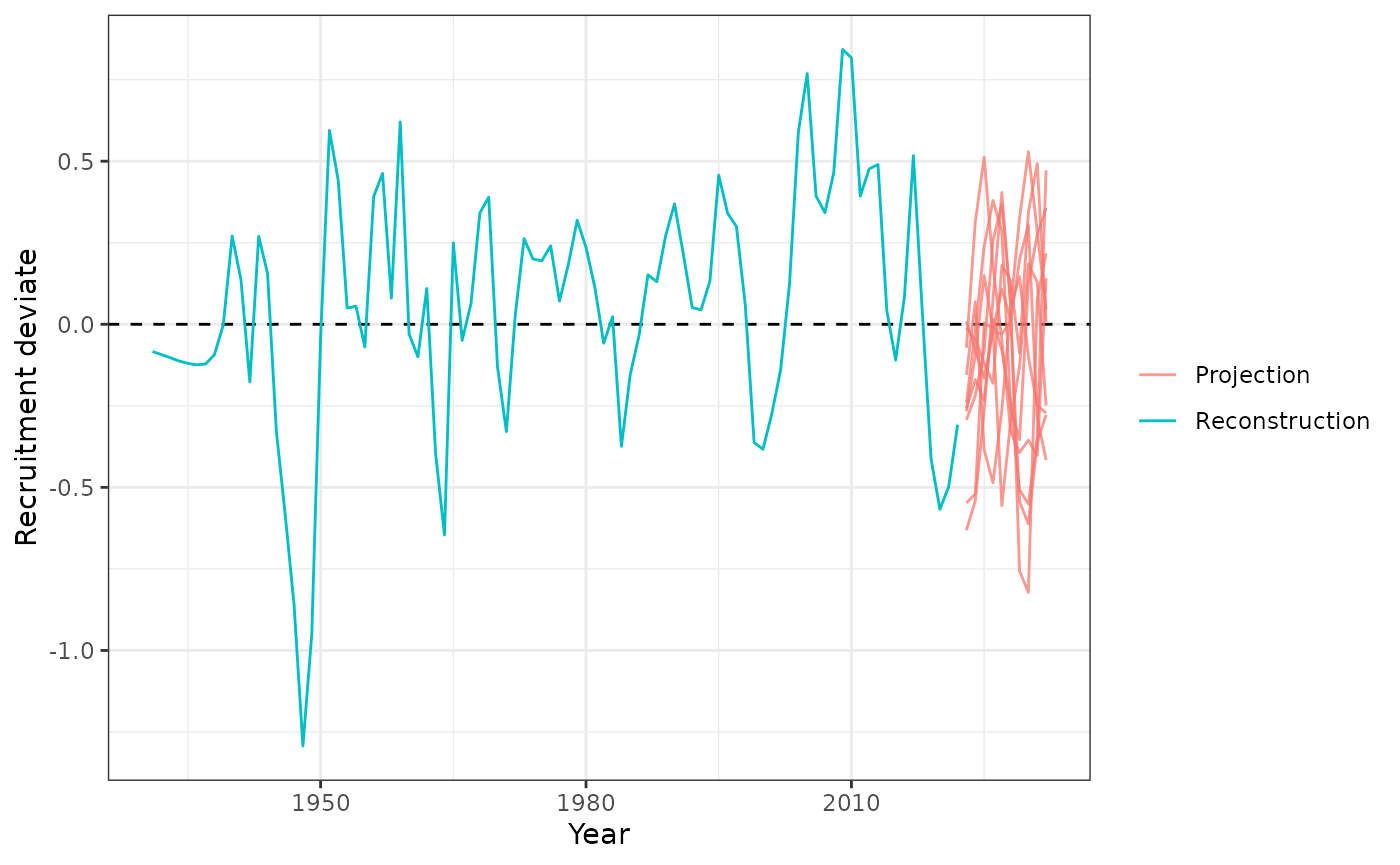
Selectivity
Projecting selectivity can be done using the
project_selectivity function. The first_yr and
the last_yr arguments define the range of years over which
the mean and standard deviation of selectivity is calculated. Future
selectivity is then drawn from a normal distribution (in log-space).
proj <- project_selectivity(
data = data, obj = obj, mcmc = NULL, first_yr = 2015, last_yr = NULL,
n_proj = n_proj, n_iter = 100)
df <- reshape2::melt(proj)
ggplot(data = df) +
geom_line(aes(x = age, y = value, color = factor(year), group = interaction(iteration, year))) +
facet_wrap(fishery ~ .)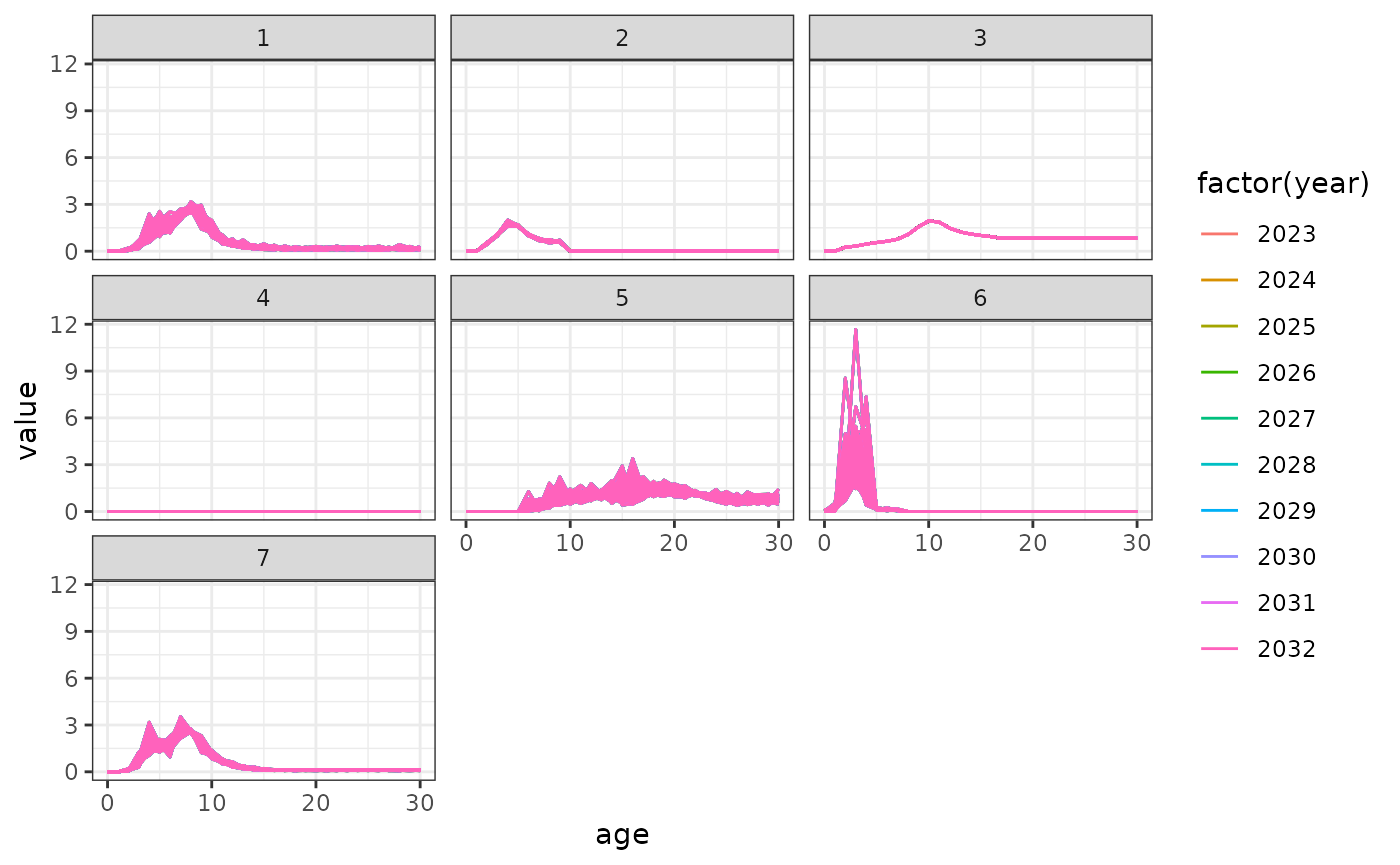
Catch
dim(data$catch_obs_ysf)
#> [1] 71 2 6
dn <- dimnames(data$catch_obs_ysf)
proj_years <- (data$last_yr + 1):(data$last_yr + n_proj)
catch_proj_ysf <- array(0, dim = list(n_proj, 2, 6), dimnames = list(Year = proj_years, Season = dn$Season, Fishery = dn$Fishery))
for (y in 1:n_proj) catch_proj_ysf[y,,] <- data$catch_obs_ysf[71,,]
catch_proj_ysf[1,,]
#> Fishery
#> Season LL1 LL2 LL3 LL4 Indonesia Australia
#> 1 0.00 0.00 1.27 0 732.54 5931.804
#> 2 9905.87 1314.67 0.00 0 0.00 0.000
plot_catch(data = data, object = obj, proj = catch_proj_ysf)
#> [1] "The maximum catch difference was: NA"
#> Warning: Removed 10 rows containing missing values or values outside the scale range
#> (`geom_line()`).